Search Count: 178
 |
Crystal Structure Sensory Appendage Protein 2 From Anopheles Culicifacies In Space Group P21 With Three Molecules Per Asu
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: LIPID BINDING PROTEIN Ligands: CD, CL, ACT |
 |
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: LIPID BINDING PROTEIN Ligands: CD, CL |
 |
Crystal Structure Of Anopheles Culicifacies Prolyl-Trna Synthetase (Acprs) In Complex With Halofuginone
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: LIGASE/INHIBITOR Ligands: HFG, ZN, CL, GOL |
 |
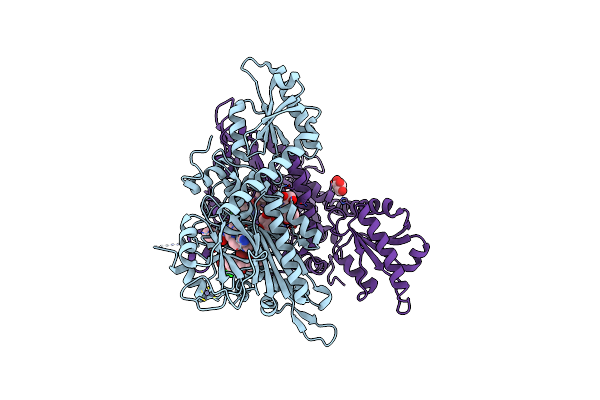
Crystal Structure Of Anopheles Culicifacies Prolyl-Trna Synthetase (Acprs) In Complex With Halofuginone And Atp Analogue
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: LIGASE/INHIBITOR Ligands: ANP, HFG, MG, ZN |
 |
Crystal Structure Of Anopheles Culicifacies Prolyl-Trna Synthetase (Acprs) In Complex With Two Inhibitors (Halofuginone And L95)
Organism: Anopheles culicifacies
Method: X-RAY DIFFRACTION Release Date: 2024-11-27 Classification: LIGASE/INHIBITOR Ligands: ZN, HFG, JE6, BR, GOL, CL |
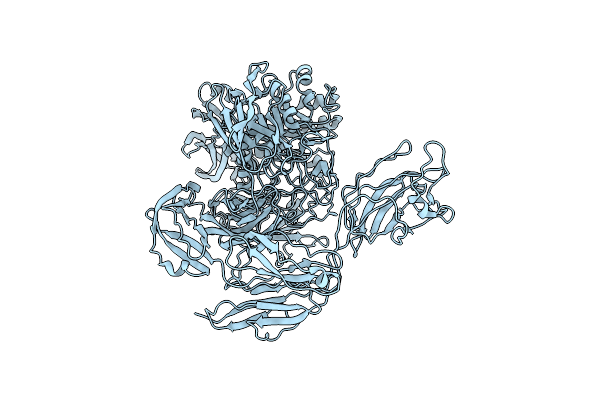 |
Organism: Niallia circulans
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: HYDROLASE |
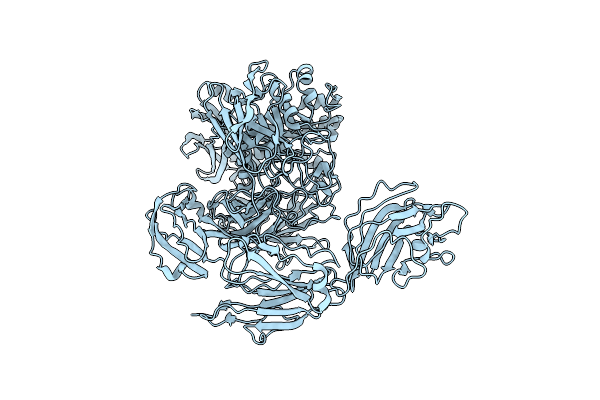 |
Organism: Niallia circulans
Method: ELECTRON MICROSCOPY Release Date: 2024-10-16 Classification: HYDROLASE |
 |
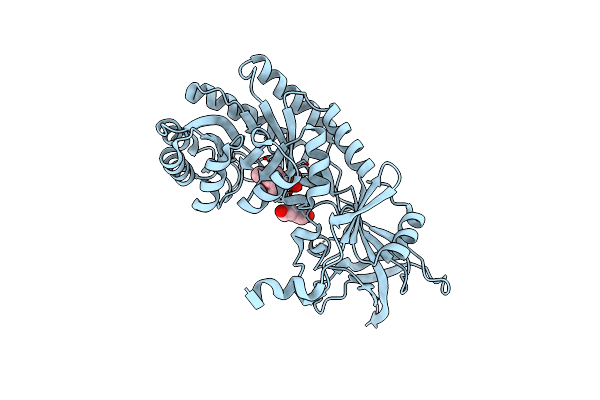
Organism: Pseudomonas phage dms3
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2024-05-08 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Btrk, A Decarboxylase Involved In Butirosin Biosynthesis
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-08-17 Classification: LYASE Ligands: PLP, PEG |
 |
Organism: Homo sapiens, Niallia circulans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-03 Classification: SIGNARING PROTEIN/INHIBITOR Ligands: EPE, GI9 |
 |
Crystal Structure Of Somatostatin Receptor 2 (Sstr2) With Peptide Antagonist Cyn 154806
Organism: Homo sapiens, Niallia circulans, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-08-03 Classification: SIGNALING PROTEIN/INHIBITOR |
 |
Organism: Homo sapiens, Bacillus circulans
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: MEMBRANE PROTEIN Ligands: 7ZQ |
 |
Organism: Homo sapiens, Bacillus circulans
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: MEMBRANE PROTEIN Ligands: 7ZQ |
 |
Organism: Homo sapiens, Bacillus circulans
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: MEMBRANE PROTEIN Ligands: GTP, MG, 7ZQ |
 |
Organism: Homo sapiens, Bacillus circulans
Method: ELECTRON MICROSCOPY Release Date: 2021-12-29 Classification: MEMBRANE PROTEIN Ligands: MG, GDP, 7ZQ |
 |
Structure Of The D125N Mutant Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6-Alpha-Manno-Cyclophellitol Carbasugar-Stabilised Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: EDO, QE8, MAN |
 |
Structure Of The D125N Mutant Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6-Alpha-Manno-Cyclophellitol Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: PBW, EDO |
 |
Structure Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6- Alpha-Manno-Cyclophellitol Carbasugar-Stabilised Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: EDO, PBW, M96, MAN |
 |
Structure Of The Catalytic Domain Of The Bacillus Circulans Alpha-1,6 Mannanase In Complex With An Alpha-1,6-Alpha-Manno-Cyclophellitol Trisaccharide Inhibitor
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2021-04-28 Classification: HYDROLASE Ligands: UKQ, UKT |
 |
Crystal Structure Of Beta-Galactosidase Ii From Bacillus Circulans In Complex With Beta-D-Galactopyranosyl Disaccharide
Organism: Bacillus circulans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-09 Classification: HYDROLASE Ligands: GLC, GAL |

