Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
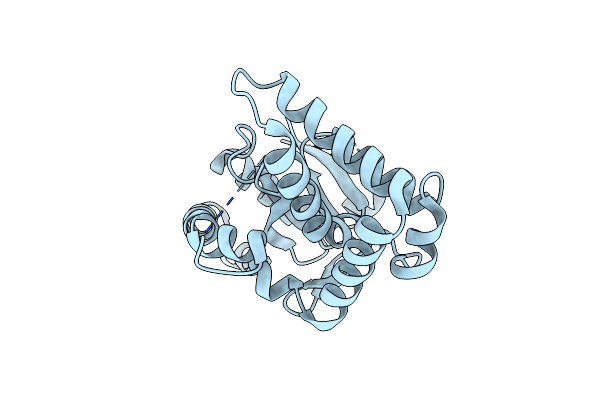 |
Crystal Structure Of A Cyclodipeptide Synthase From Parcubacteria Bacterium Raac4_Od1_1, E174A Mutant
Organism: Parcubacteria bacterium raac4_od1_1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-09-28 Classification: LIGASE |
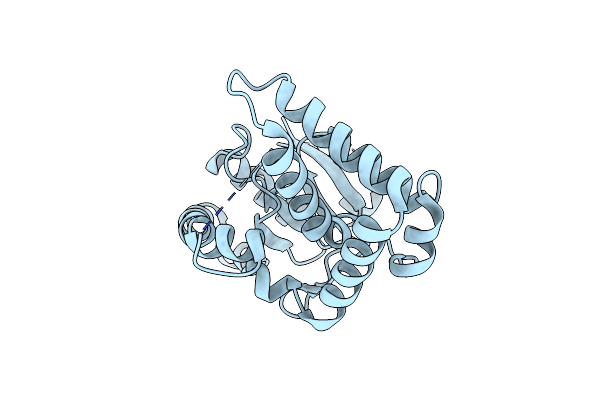 |
Crystal Structure Of A Cyclodipeptide Synthase From Parcubacteria Bacterium Raac4_Od1_1, E174L
Organism: Parcubacteria bacterium raac4_od1_1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-09-28 Classification: LIGASE |
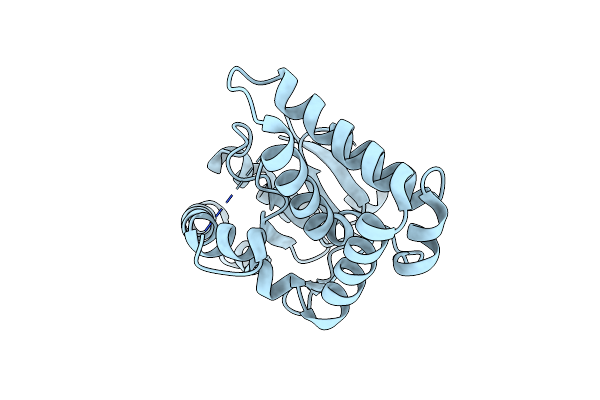 |
Crystal Structure Of A Cyclodipeptide Synthase From Parcubacteria Bacterium Raac4_Od1_1, D58N Mutant
Organism: Parcubacteria bacterium raac4_od1_1
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-09-28 Classification: LIGASE |
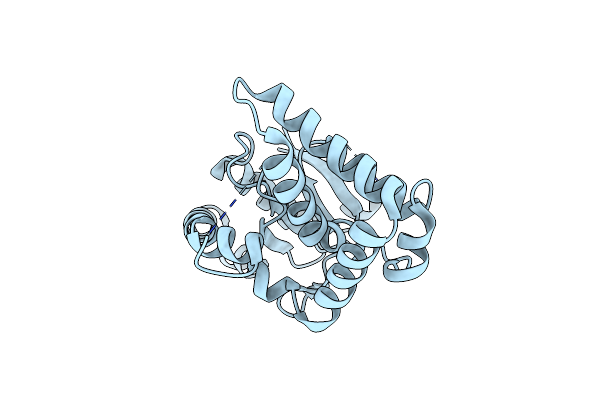 |
Crystal Structure Of A Cyclodipeptide Synthase From Parcubacteria Bacterium Raac4_Od1_1, Y189F Mutant
Organism: Parcubacteria bacterium raac4_od1_1
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2022-09-28 Classification: LIGASE |
 |
Crystal Structure Of A Cyclodipeptide Synthase From Parcubacteria Bacterium Raac4_Od1_1, E171Q Mutant
Organism: Parcubacteria bacterium raac4_od1_1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-09-28 Classification: LIGASE |
 |
Crystal Structure Of A Cyclodipeptide Synthase From Parcubacteria Bacterium Raac4_Od1_1, Y55F Mutant
Organism: Parcubacteria bacterium raac4_od1_1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-09-28 Classification: LIGASE |
 |
Crystal Structure Of A Cyclodipeptide Synthase From Parcubacteria Bacterium Raac4_Od1_1, Wt Form
Organism: Parcubacteria bacterium raac4_od1_1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-09-28 Classification: LIGASE |
 |
Crystal Structure Of Triosephosphate Isomerase From Candidatus Absconditabacteria (Sr1) Bacterium
Organism: Candidate division sr1 bacterium raac1_sr1_1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-07-20 Classification: ISOMERASE |
 |
Crystal Structure Of Triosephosphate Isomerase From Candidate Division Katanobacteria (Wwe3) Bacterium
Organism: Candidate division wwe3 bacterium raac2_wwe3_1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: ISOMERASE Ligands: PO4 |
 |
Crystal Structure Of E36-G37Del Mutant Of The Bacillus Caldolyticus Cold Shock Protein.
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-05-10 Classification: DNA BINDING PROTEIN Ligands: SO4 |
 |
Crystal Structure Of Pyrr, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus Caldolyticus, Ump-Bound Form
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2007-09-25 Classification: TRANSFERASE Ligands: U5P, EDO |
 |
Organism: Eimeria tenella
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-09-18 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Bacillus Caldolyticus Cold Shock Protein In Complex With Hexathymidine
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:1.29 Å Release Date: 2007-04-24 Classification: GENE REGULATION/DNA Ligands: CA, MPD |
 |
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2006-01-03 Classification: GENE REGULATION Ligands: ACY |
 |
Pyrr, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus Caldolyticus, Nucleotide-Bound Form
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2005-03-01 Classification: TRANSCRIPTION, TRANSFERASE Ligands: MG, 5GP, 3GP, U5P |
 |
Pyrr, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus Caldolyticus, Sulfate-Bound Form
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2005-03-01 Classification: TRANSCRIPTION, TRANSFERASE Ligands: MG, SO4 |
 |
Pyrr, The Regulator Of The Pyrimidine Biosynthetic Operon In Bacillus Caldolyticus
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-05-25 Classification: TRANSCRIPTION, TRANSFERASE |
 |
Crystal Structure Of Bacillus Caldolyticus Uracil Phosphoribosyltransferase With Bound Ump
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2002-06-05 Classification: TRANSFERASE Ligands: U5P |
 |
Bacillus Caldolyticus Cold-Shock Protein Mutants To Study Determinants Of Protein Stability
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-11-07 Classification: TRANSCRIPTION |
 |
Bacillus Caldolyticus Cold-Shock Protein Mutants To Study Determinants Of Protein Stability
Organism: Bacillus caldolyticus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-11-07 Classification: TRANSCRIPTION |