Search Count: 59
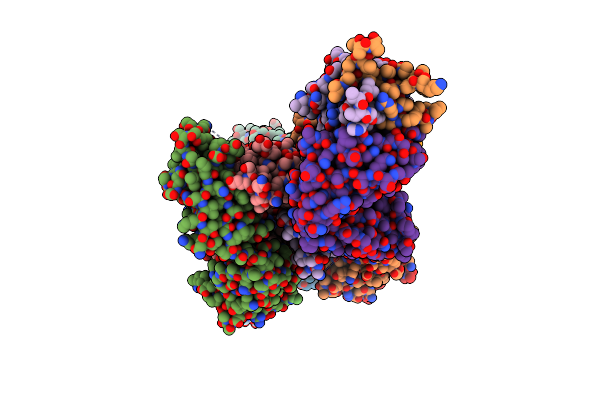 |
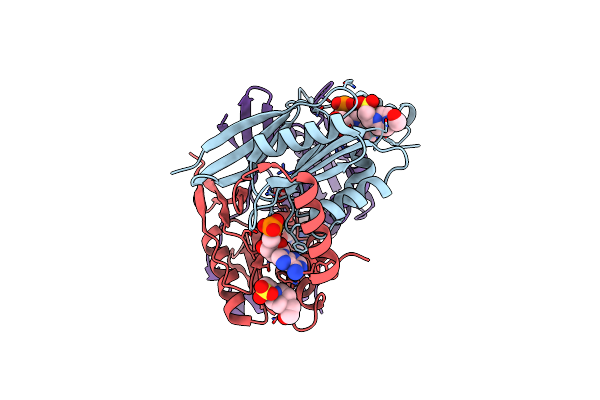
Crystal Structure Of Hepatitis C Virus E1 Glycoprotein Epitope 314-324 Scaffold Design 1W4K_08 In Complex With Neutralizing Antibody F(Ab) Fragment Igh526
Organism: Pyrobaculum aerophilum, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-10-09 Classification: STRUCTURAL PROTEIN |
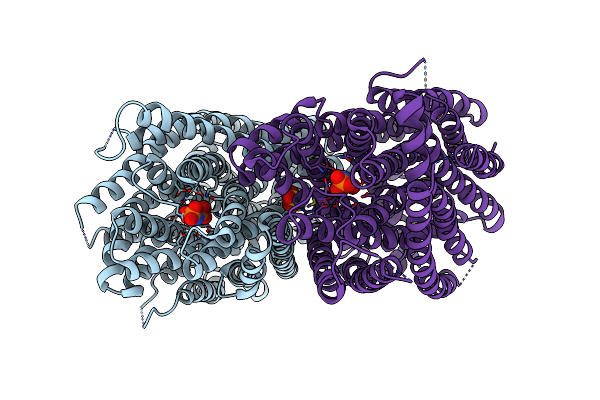 |
Crystal Structure Of Pyrobaculum Aerophilum Potassium-Independent Proton Pumping Membrane Integral Pyrophosphatase In Complex With Imidodiphosphate And Magnesium, And With Bound Sulphate
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: MG, 2PN, SO4 |
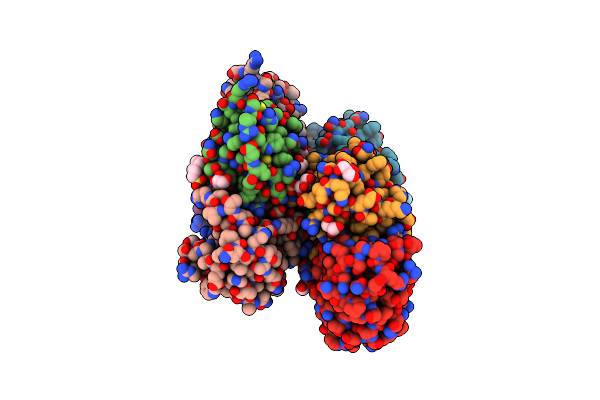 |
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2013-08-21 Classification: HYDROLASE/DE NOVO PROTEIN Ligands: PO4, GOL, EPE, SO4 |
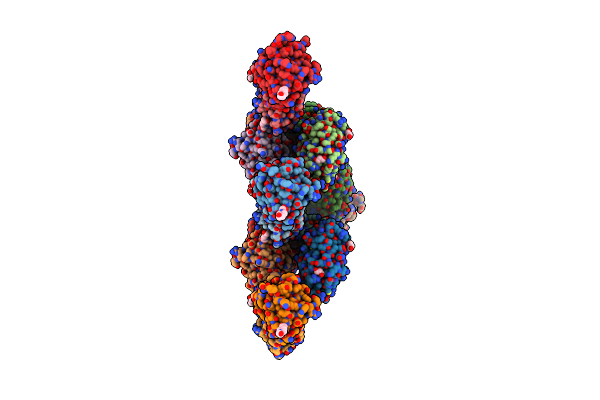 |
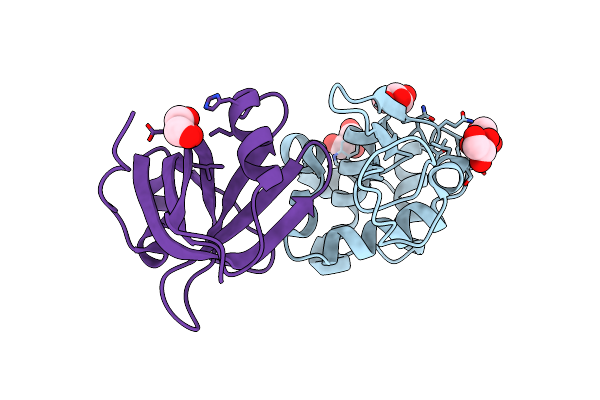
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-08-21 Classification: HYDROLASE/DE NOVO PROTEIN Ligands: GOL, EPE |
 |
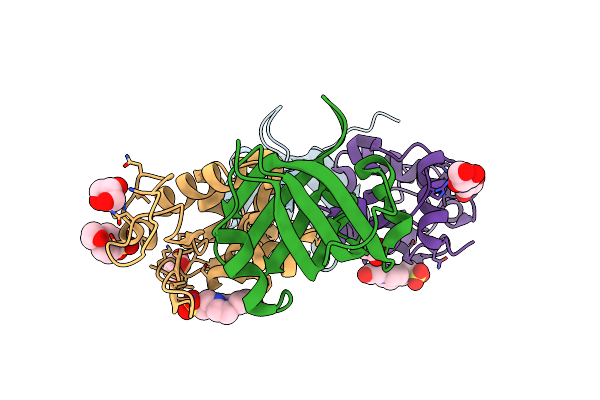
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2013-08-21 Classification: DE NOVO PROTEIN/HYDROLASE Ligands: GOL |
 |
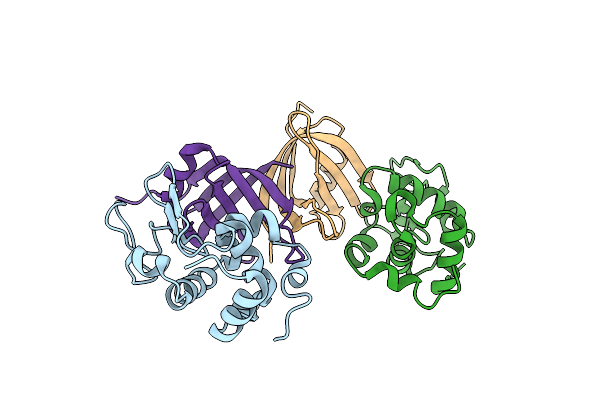
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2013-08-21 Classification: DE NOVO PROTEIN/HYDROLASE Ligands: EPE, GOL, ACT |
 |
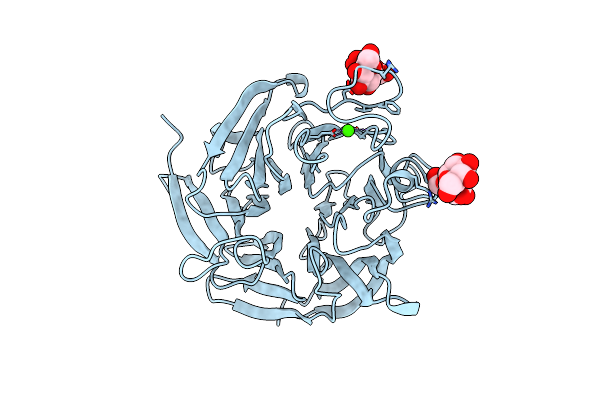
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2013-08-14 Classification: HYDROLASE/DE NOVO PROTEIN |
 |
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2010-09-08 Classification: OXIDOREDUCTASE Ligands: CA |
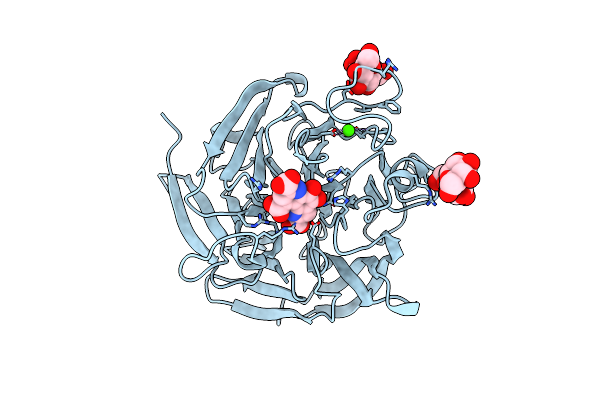 |
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-09-08 Classification: OXIDOREDUCTASE Ligands: CA, PQQ |
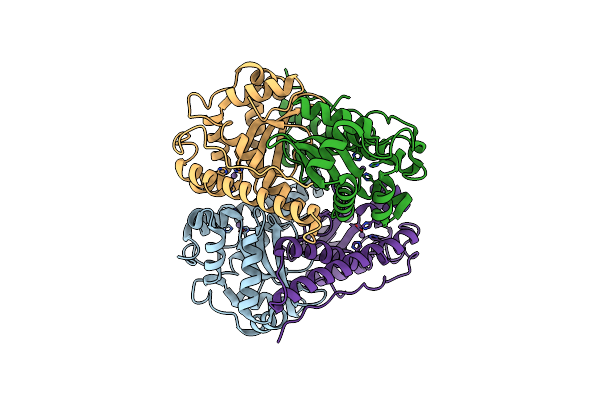 |
Crystal Structure Of The Metal-Bound Superoxide Dismutase From Pyrobaculum Aerophilum
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2009-08-25 Classification: OXIDOREDUCTASE Ligands: MN |
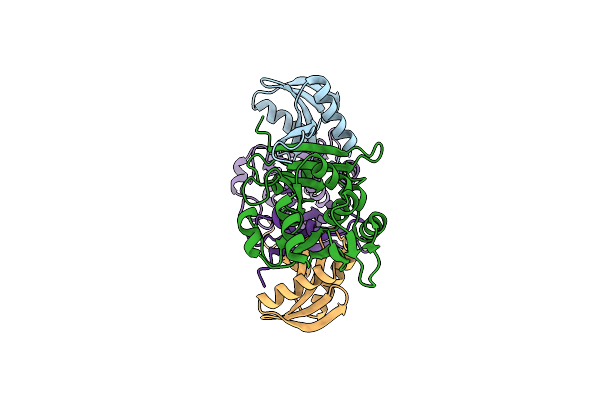 |
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2009-06-23 Classification: SPLICING |
 |
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2008-08-05 Classification: UNKNOWN FUNCTION Ligands: AMP, EPE, ACT |
 |
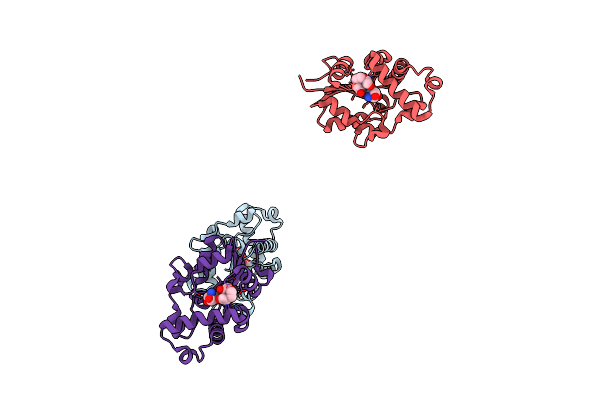
Structure Of Conserved Hypothetical Protein Pae2307 From Pyrobaculum Aerophilum
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2006-01-10 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: PO4 |
 |
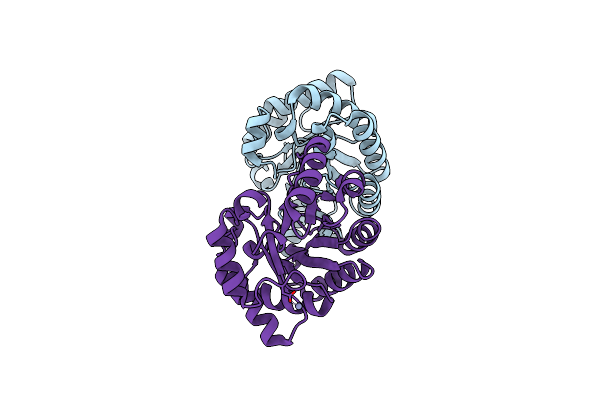
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2005-09-20 Classification: TRANSFERASE Ligands: TRS, PGE |
 |
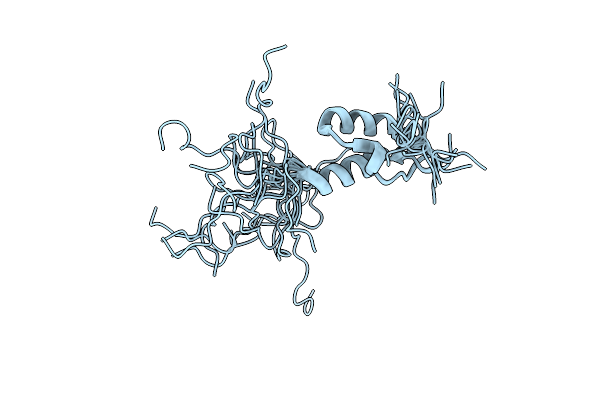
Structure Of 2-Deoxyribose-5-Phosphate Aldolase From Pyrobaculum Aerophilum
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-08-02 Classification: LYASE Ligands: ZN |
 |
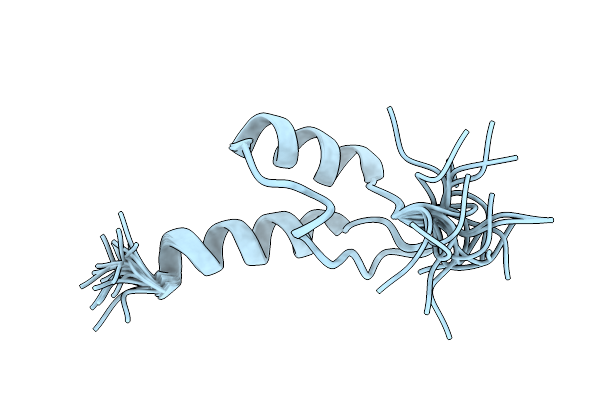
Peripheral-Subunit Binding Domains From Mesophilic, Thermophilic, And Hyperthermophilic Bacteria Fold By Ultrafast, Apparently Two-State Transitions
Organism: Pyrobaculum aerophilum
Method: SOLUTION NMR Release Date: 2005-07-20 Classification: TRANSFERASE |
 |
Peripheral-Subunit Binding Domains From Mesophilic, Thermophilic, And Hyperthermophilic Bacteria Fold By Ultrafast, Apparently Two-State Transitions
Organism: Pyrobaculum aerophilum
Method: SOLUTION NMR Release Date: 2005-07-20 Classification: TRANSFERASE |
 |
Peripheral-Subunit Binding Domains From Mesophilic, Thermophilic, And Hyperthermophilic Bacteria Fold By Ultrafast, Apparently Two-State Transitions
Organism: Pyrobaculum aerophilum
Method: SOLUTION NMR Release Date: 2005-07-20 Classification: TRANSFERASE |
 |
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2005-01-18 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: SO4, ACT, CL, P6G, PG4 |
 |
Crystal Structure Of Phosphoglucose/Phosphomannose Isomerase From Pyrobaculum Aerophilum In Complex With Fructose 6-Phosphate
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2004-12-07 Classification: ISOMERASE Ligands: SO4, F6R, GOL |

