Search Count: 334
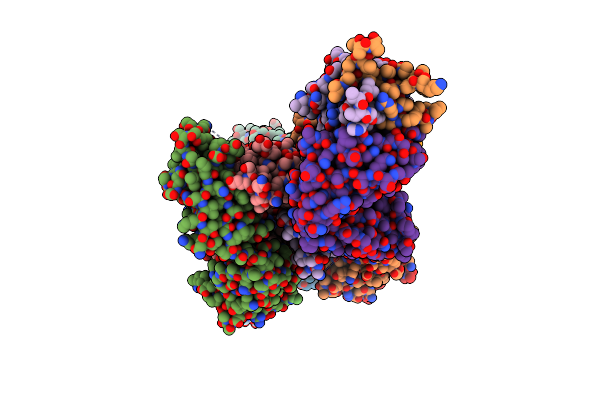 |
Crystal Structure Of Hepatitis C Virus E1 Glycoprotein Epitope 314-324 Scaffold Design 1W4K_08 In Complex With Neutralizing Antibody F(Ab) Fragment Igh526
Organism: Pyrobaculum aerophilum, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-10-09 Classification: STRUCTURAL PROTEIN |
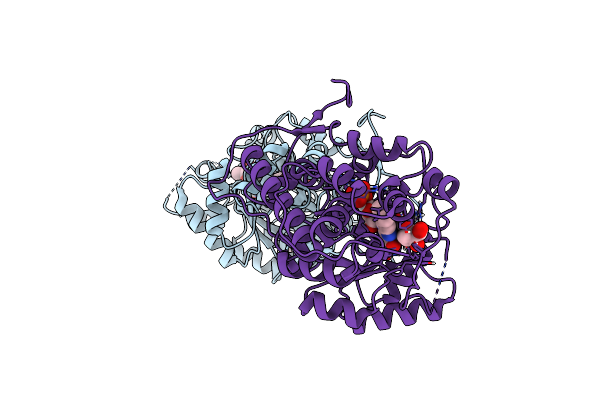 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-08-14 Classification: FLAVOPROTEIN Ligands: FMN, EDO, LAC, PYR |
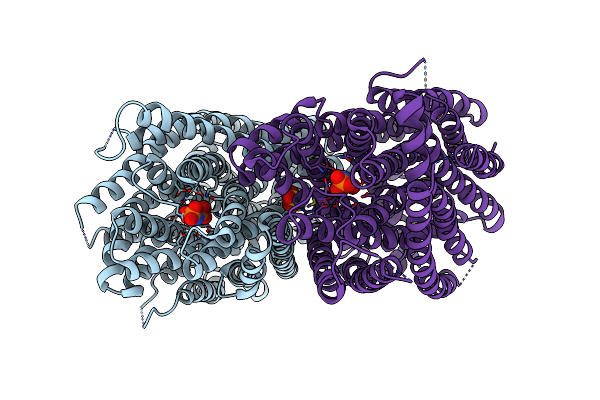 |
Crystal Structure Of Pyrobaculum Aerophilum Potassium-Independent Proton Pumping Membrane Integral Pyrophosphatase In Complex With Imidodiphosphate And Magnesium, And With Bound Sulphate
Organism: Pyrobaculum aerophilum
Method: X-RAY DIFFRACTION Resolution:3.84 Å Release Date: 2024-01-17 Classification: MEMBRANE PROTEIN Ligands: MG, 2PN, SO4 |
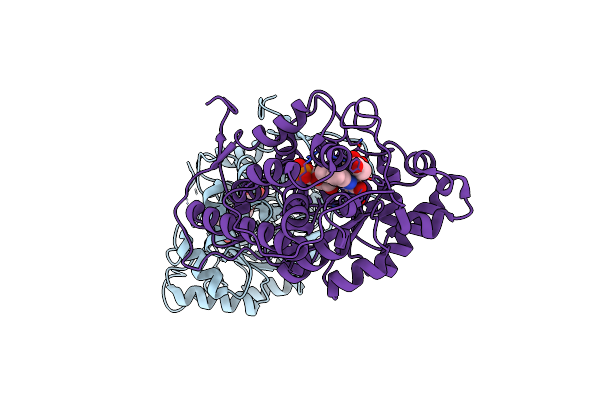 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2022-06-15 Classification: OXIDOREDUCTASE Ligands: ACT, FMN, EDO |
 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2022-03-23 Classification: OXIDOREDUCTASE Ligands: FMN, 2OP |
 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2022-03-23 Classification: OXIDOREDUCTASE Ligands: FMN, LAC |
 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-03-23 Classification: OXIDOREDUCTASE Ligands: FMN, PYR |
 |
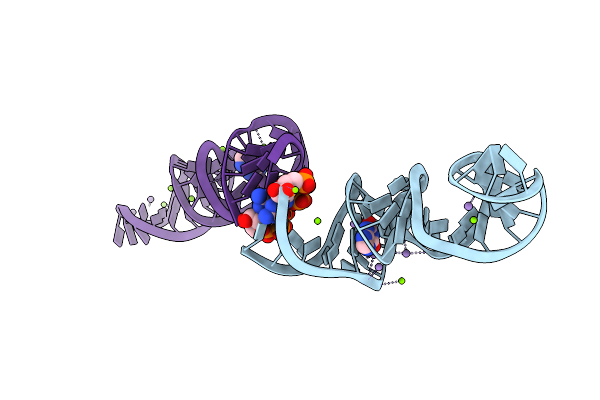
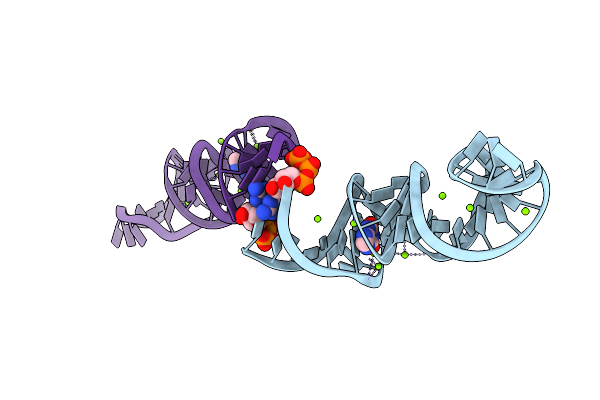
Crystal Structure Of Xanthine Riboswitch With Xanthine, Iridium Hexammine Soak
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, IR, XAN, MG |
 |
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, XAN, MN, MG |
 |
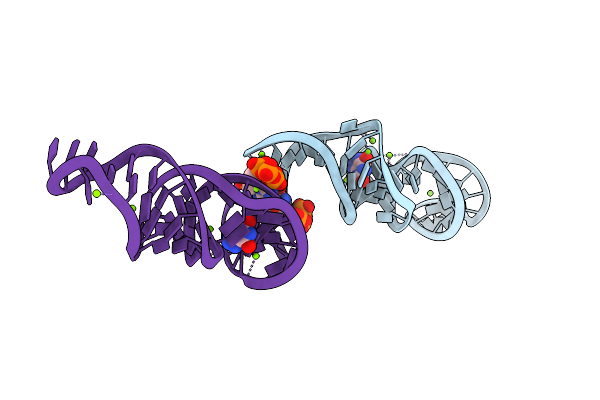
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, XAN, MG |
 |
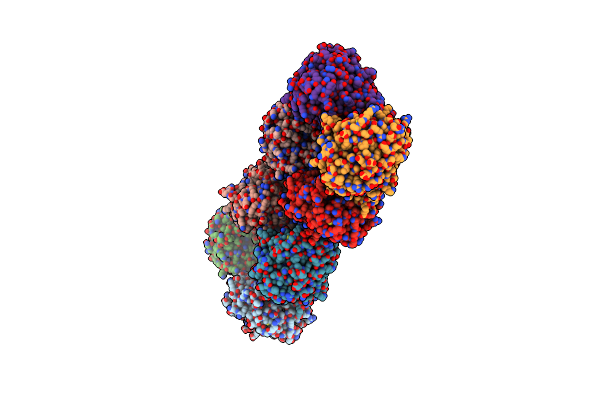
Organism: Ideonella sp. b508-1
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-06-30 Classification: RNA Ligands: GTP, MG, AZA |
 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-06-29 Classification: OXIDOREDUCTASE Ligands: FMN, PYR |
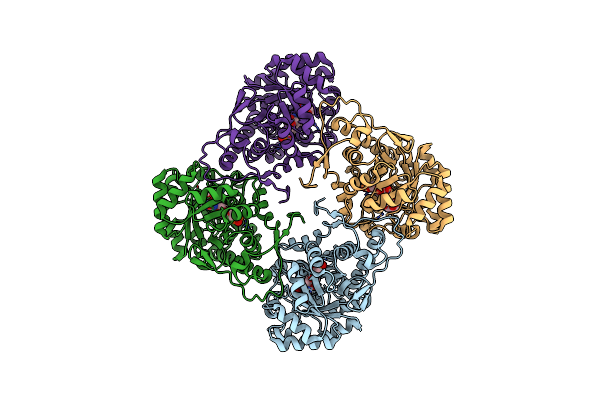 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-08-26 Classification: OXIDOREDUCTASE Ligands: FMN, PYR |
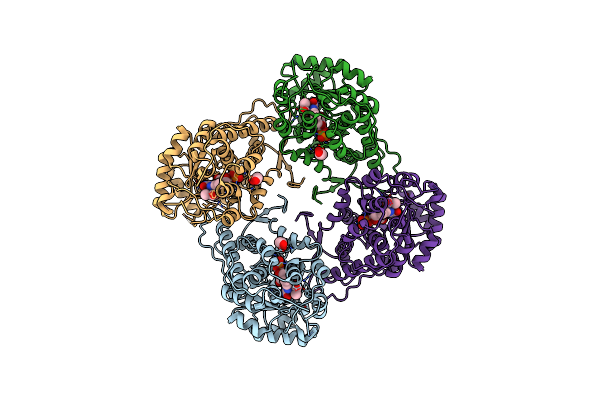 |
Organism: Aerococcus viridans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-12-03 Classification: OXIDOREDUCTASE Ligands: FNR, PYR, EDO |
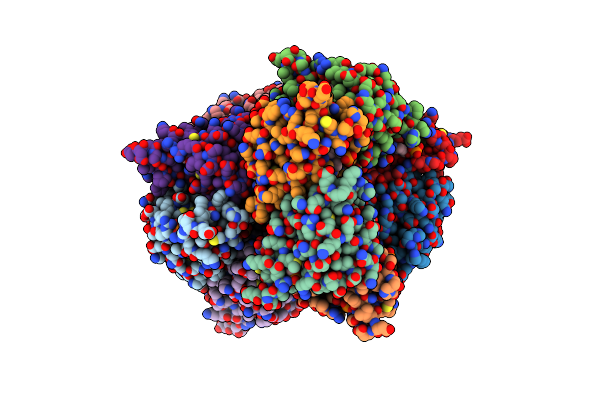 |
Crystal Structure Of Dcmp Deaminase From The Cyanophage S-Tim5 In Complex With Dcmp And Dump
Organism: Cyanophage s-tim5
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-11-26 Classification: HYDROLASE Ligands: ZN, DCM, DU |
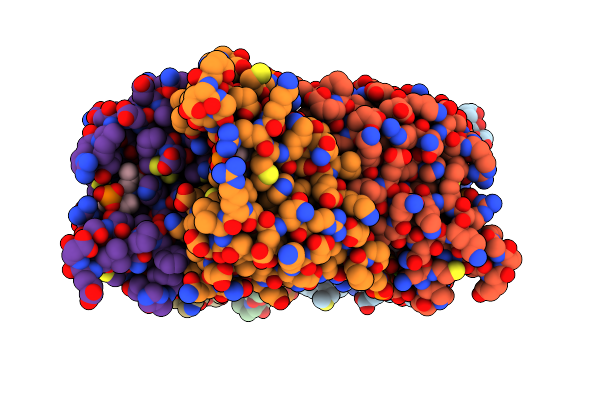 |
Crystal Structure Of Dcmp Deaminase From The Cyanophage S-Tim5 In Complex With Dtmp And Dttp.
Organism: Cyanophage s-tim5
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2014-11-26 Classification: HYDROLASE Ligands: ZN, MG, TTP, TMP, CL |
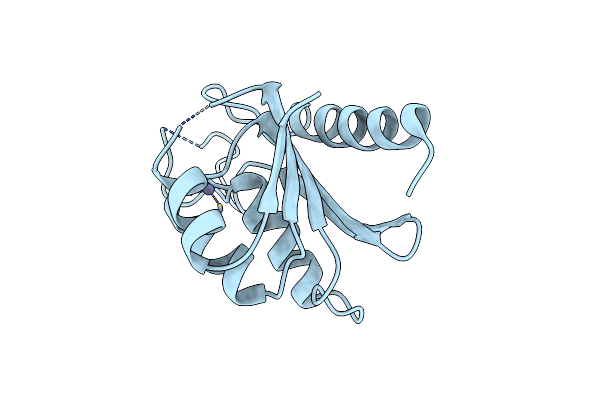 |
Organism: Cyanophage s-tim5
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-11-26 Classification: HYDROLASE Ligands: ZN, CL |
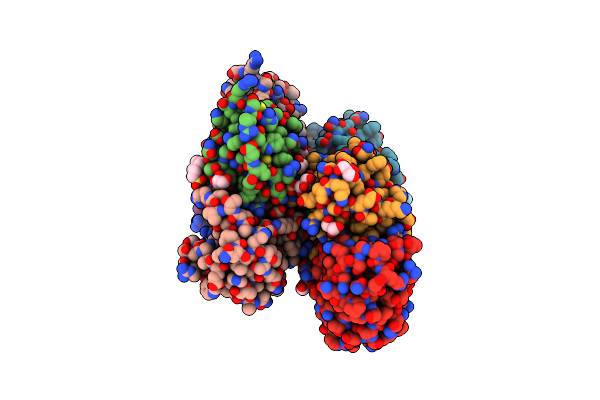 |
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2013-08-21 Classification: HYDROLASE/DE NOVO PROTEIN Ligands: PO4, GOL, EPE, SO4 |
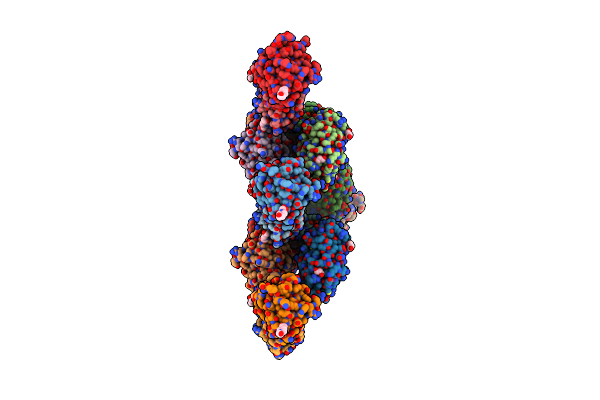 |
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-08-21 Classification: HYDROLASE/DE NOVO PROTEIN Ligands: GOL, EPE |
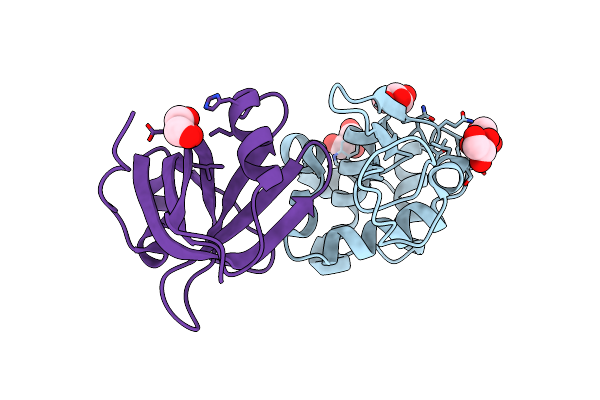 |
Organism: Pyrobaculum aerophilum, Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2013-08-21 Classification: DE NOVO PROTEIN/HYDROLASE Ligands: GOL |

