Search Count: 191
 |
Nmr Solution Structure Of Termini-Truncated Oxidized Cytochrome C552 From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: SOLUTION NMR Release Date: 2025-07-30 Classification: ELECTRON TRANSPORT Ligands: HEC |
 |
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: CU, ACT, SO4 |
 |
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-04-24 Classification: METAL BINDING PROTEIN Ligands: PEG, CU |
 |
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-04-03 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
Exploring The Ligand Binding And Conformational Dynamics Of Receptor Domain 1 Of The Abc Transporter Glnpq
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-10-04 Classification: TRANSPORT PROTEIN Ligands: MPD, GOL, GLN, NA, PEG |
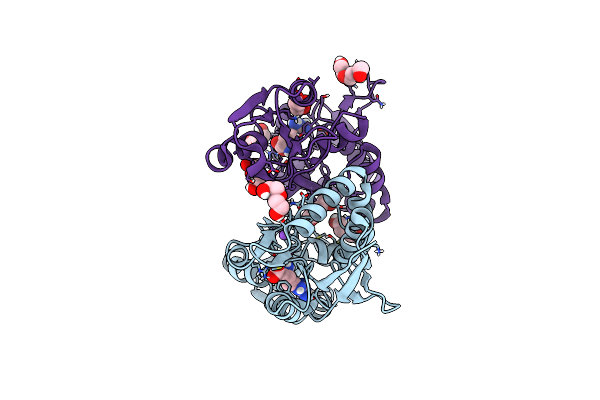 |
Exploring The Ligand Binding And Conformational Dynamics Of Receptor Domain 1 Of The Abc Transporter Glnpq
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-10-04 Classification: TRANSPORT PROTEIN Ligands: ARG, NA, GOL, PEG, PGE |
 |
The Structure Of Thiocyanate Dehydrogenase Mutant Form With Lys 281 Replaced By Ala From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-06-07 Classification: OXIDOREDUCTASE Ligands: SO4, BO3, CU, NA |
 |
The Structure Of Thiocyanate Dehydrogenase Mutant Form With Thr 169 Replaced By Ala From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-05-31 Classification: OXIDOREDUCTASE Ligands: CU, SO4 |
 |
The Structure Of Thiocyanate Dehydrogenase Mutant Form With Phe 436 Replaced By Gln From Thioalkalivibrio Paradoxus
Organism: Thioalkalivibrio paradoxus arh 1
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2023-01-18 Classification: OXIDOREDUCTASE Ligands: PEG, CU |
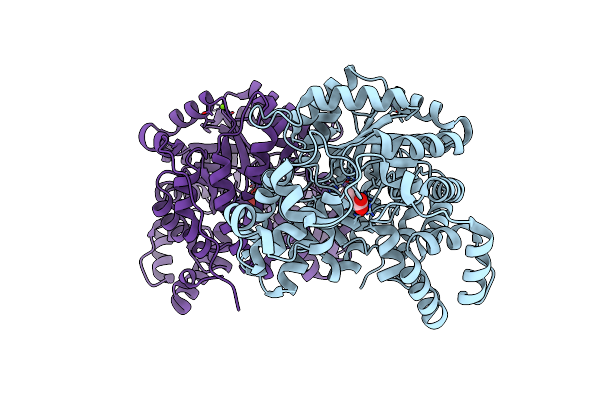 |
Cryo-Em Structure Of "Ct-Ct Dimer" Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: LIGASE Ligands: MG, MN, PYR |
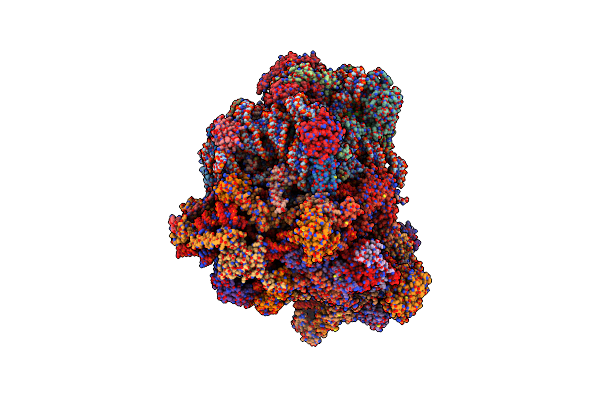 |
Organism: Spraguea lophii 42_110
Method: ELECTRON MICROSCOPY Resolution:2.79 Å Release Date: 2022-11-30 Classification: RIBOSOME Ligands: K, MG, ZN |
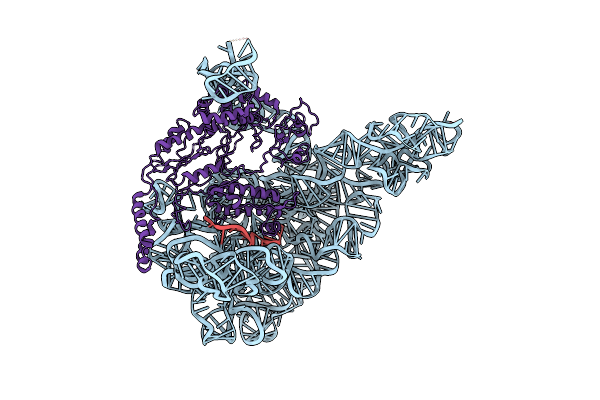 |
Cryo-Em Structure Of A Group Ii Intron Complexed With Its Reverse Transcriptase
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: RNA BINDING PROTEIN/RNA |
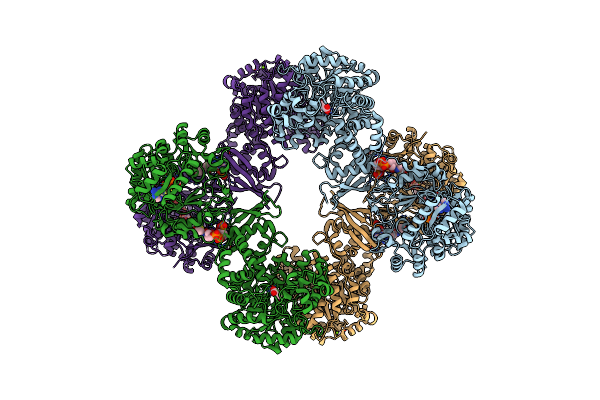 |
Cryo-Em Structure Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN, ADP, ACO, PYR, BCT |
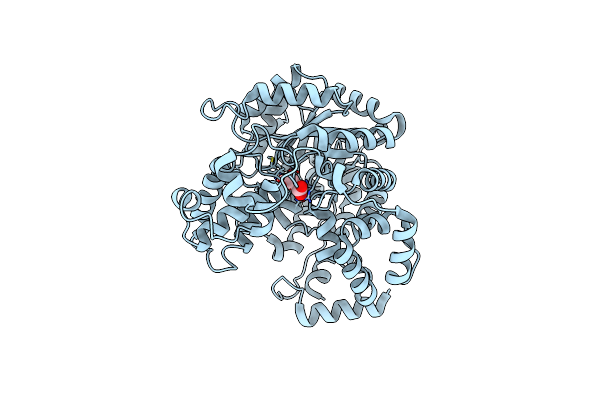 |
Cryo-Em Structure Of "Ct Oxa" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MN, OAA |
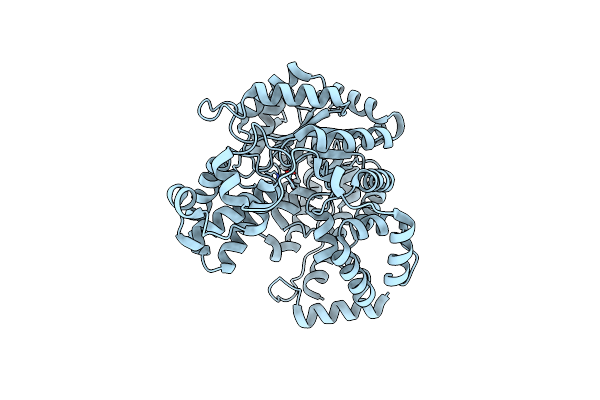 |
Cryo-Em Structure Of "Ct Empty" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN |
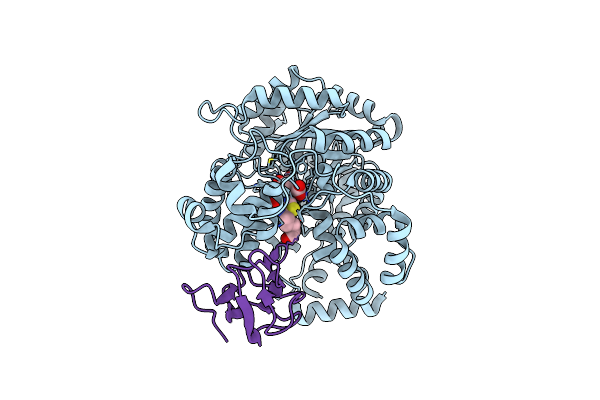 |
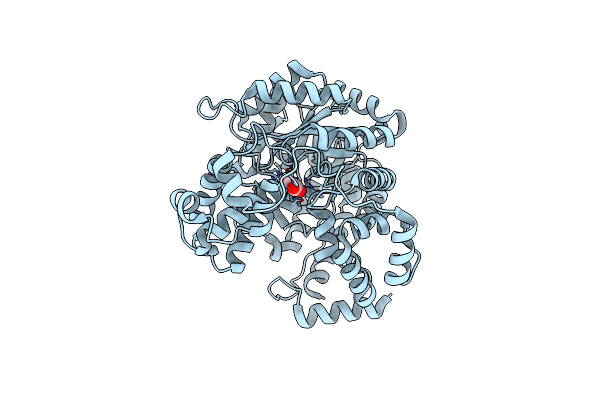
Cryo-Em Structure Of "Ct React" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN, OAA, BTN |
 |
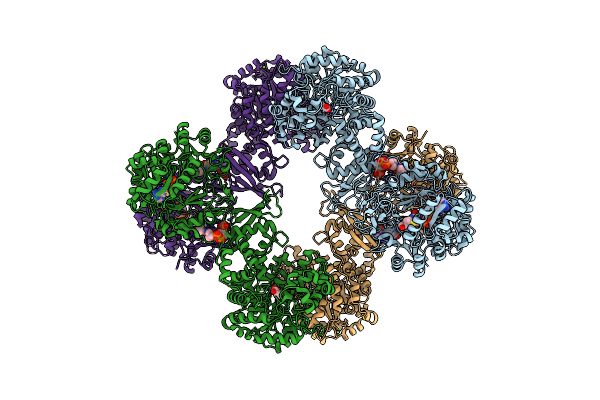
Cryo-Em Structure Of "Ct Pyr" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: MG, MN, PYR |
 |
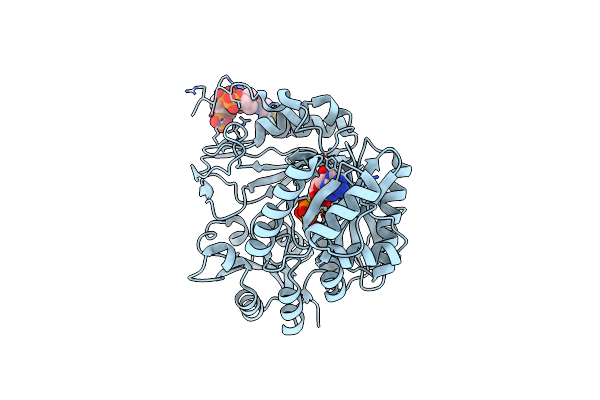
Cryo-Em Structure Of "Bc React" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: BTN, MG, MN, ATP, ACO, PYR, BCT, ADP |
 |
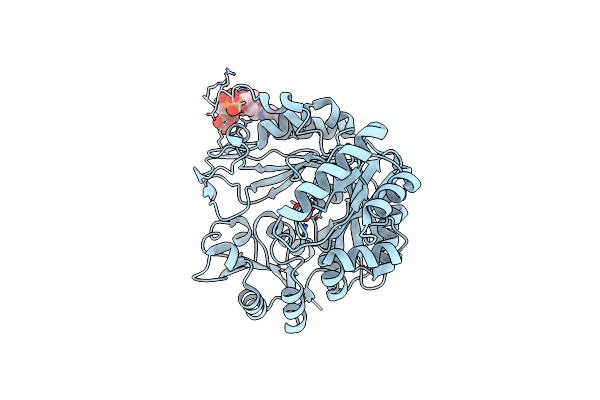
Cryo-Em Structure Of "Bc Closed" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: ATP, MG, ACO |
 |
Cryo-Em Structure Of "Bc Open" Conformation Of Lactococcus Lactis Pyruvate Carboxylase With Acetyl-Coa
Organism: Lactococcus lactis
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: LIGASE Ligands: ACO, BCT |

