Search Count: 219
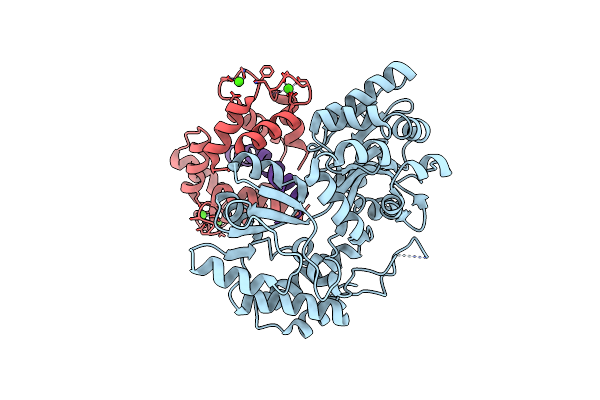 |
Organism: Escherichia coli #1/h766, Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-04-02 Classification: PROTEIN BINDING Ligands: CA |
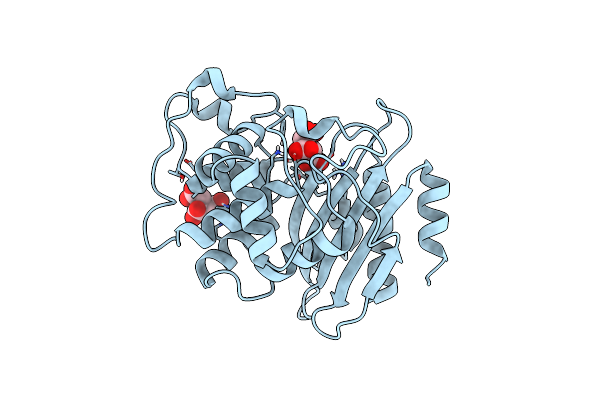 |
Crystal Structure Of A Class A Beta-Lactamase From Nocardia Cyriacigeorgica
Organism: Nocardia cyriacigeorgica
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-01-17 Classification: HYDROLASE Ligands: FLC |
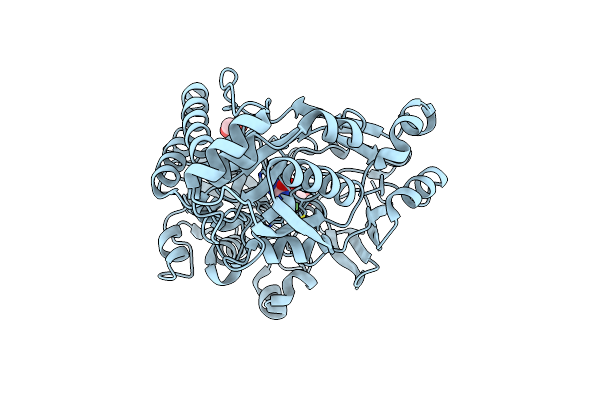 |
Structure Of A Periplasmic Peptide Binding Protein From Mesorhizobium Sp. Ap09 Bound To Aminoserine
Organism: Mesorhizobium sp. ap09
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-11-08 Classification: PEPTIDE BINDING PROTEIN Ligands: CA, EDO, SET |
 |
Organism: Xenorhabdus poinarii g6
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-09-21 Classification: CELL CYCLE Ligands: ADP, MG |
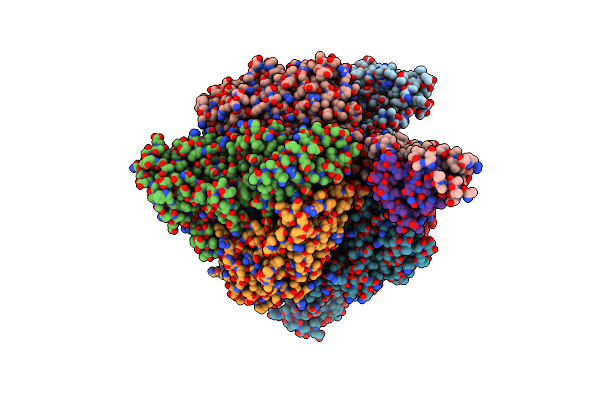 |
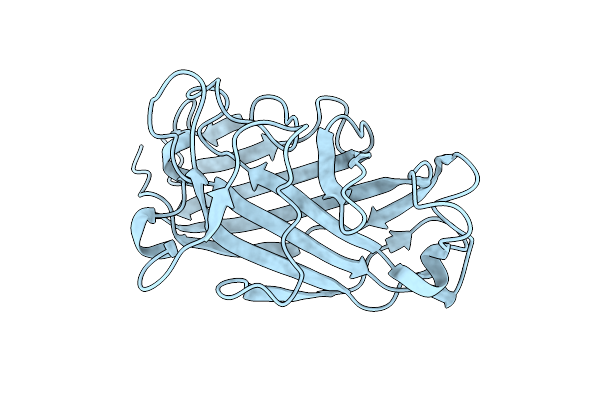
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:3.82 Å Release Date: 2022-06-22 Classification: MOTOR PROTEIN Ligands: GOL, ADP, MG, ALF |
 |
Structure Of The C-Terminal Head Domain Of The Fowl Adenovirus Type 4 Fiber 1
Organism: Fowl adenovirus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-03-16 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Marr Family Transcriptional Regulator From Enterobacter Soli Strain Lf7 Bound To Indole 3 Acetic Acid
Organism: Enterobacter soli atcc baa-2102
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2021-12-22 Classification: DNA BINDING PROTEIN Ligands: IAC, NI |
 |
Crystal Structure Of The Light-Harvesting Complex Ii (B800-850) From Ectothiorhodospira Haloalkaliphila
Organism: Ectothiorhodospira haloalkaliphila, Halorhodospira halochloris str. a
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2019-10-16 Classification: PHOTOSYNTHESIS Ligands: BCL, LYC, DET |
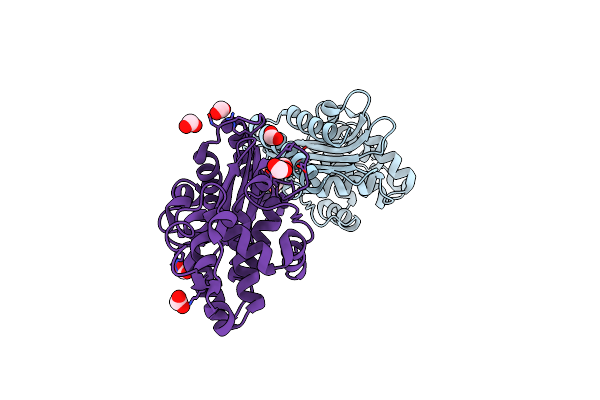 |
Crystal Structure Of The Beta Lactamase Class A Penp From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-01-30 Classification: HYDROLASE Ligands: EDO, FMT |
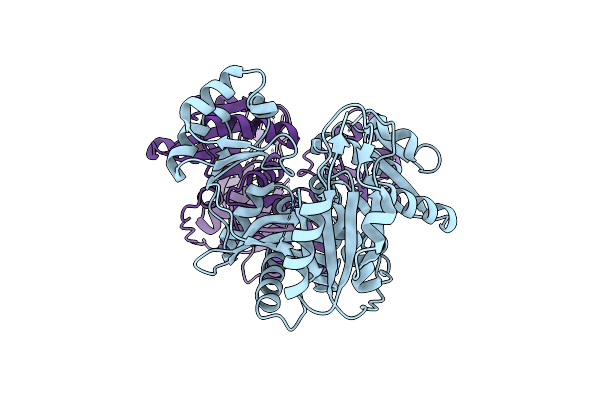 |
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-09-09 Classification: LIGASE |
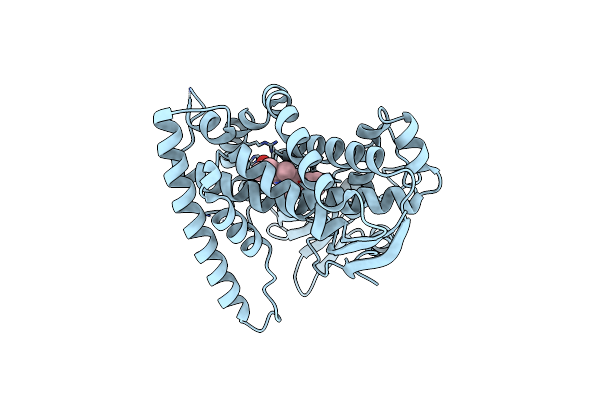 |
The Crystal Structure Of The Versatile Cytochrome P450 Enzyme Cyp109B1 From Bacillus Subtilis
Organism: Bacillus subtilis subsp. subtilis
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2015-02-04 Classification: ELECTRON TRANSPORT Ligands: HEM |
 |
Organism: Influenza b virus
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2014-04-02 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Nucleotide-Free A3B3 Complex From Enterococcus Hirae V-Atpase [Ea3B3]
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-01-16 Classification: HYDROLASE |
 |
Crystal Structure Of Amp-Pnp Bound A3B3 Complex From Enterococcus Hirae V-Atpase [Ba3B3]
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: ANP, MG |
 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: GOL, CL, B3P |
 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:3.90 Å Release Date: 2013-01-16 Classification: HYDROLASE |
 |
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2013-01-16 Classification: HYDROLASE Ligands: ANP, MG |
 |
Crystal Structure Of The Central Axis (Ntpd-Ntpg) In The Catalytic Portion Of Enterococcus Hirae V-Type Sodium Atpase
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-10-05 Classification: HYDROLASE Ligands: NO3 |
 |
Structure Of The Na+ Unbound Rotor Ring Modified With N,N F-Dicyclohexylcarbodiimide Of The Na+-Transporting V-Atpase
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:3.14 Å Release Date: 2011-08-17 Classification: HYDROLASE Ligands: DCW, UMQ |
 |
Crystal Structure Of Rotor Ring With Dccd Of The V- Atpase From Enterococcus Hirae
Organism: Enterococcus hirae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-12-05 Classification: HYDROLASE Ligands: DCW, NA, LHG, UMQ |

