Search Count: 117
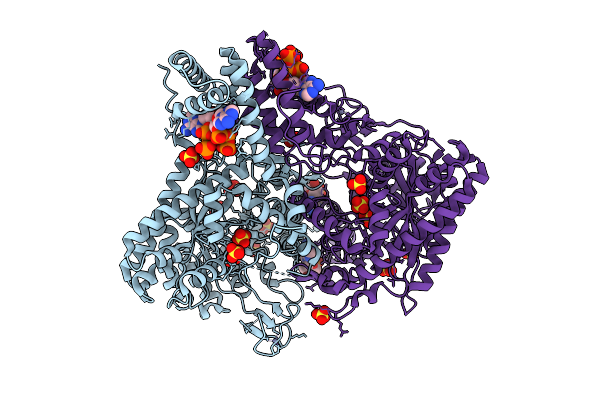 |
Crystal Structure Of S. Thermophilus Class Iii Ribonucleotide Reductase Bound To Datp
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: MG, DTP, ZN, SO4 |
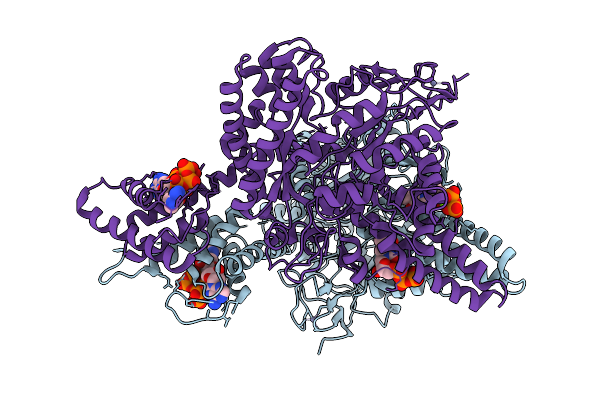 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: TTP, DTP, MG, ZN |
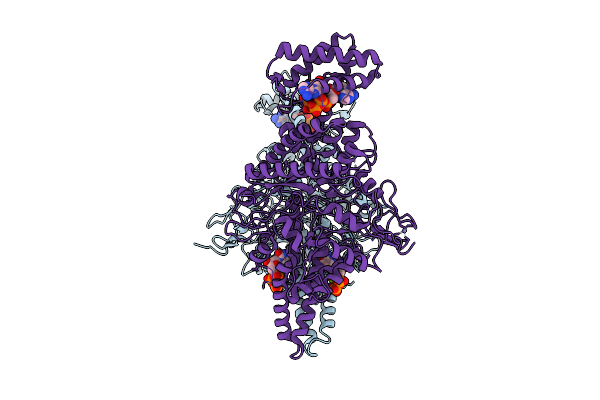 |
Organism: Streptococcus thermophilus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: ATP, MG, TTP, ZN |
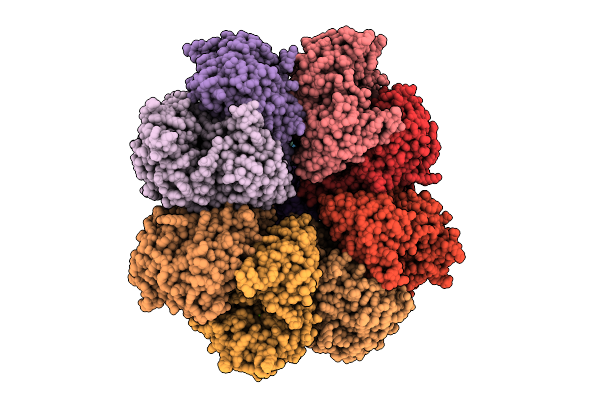 |
Organism: Streptococcus thermophilus, Streptococcus phage 2972
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
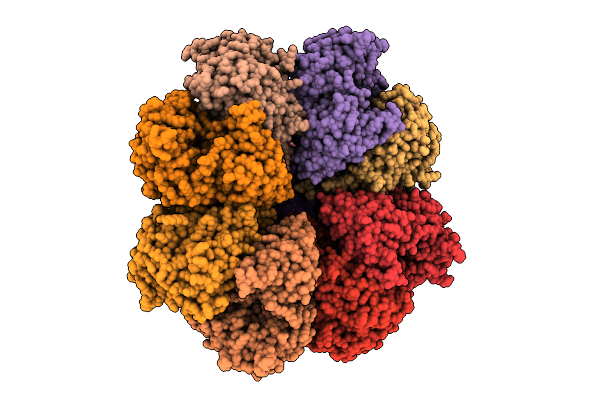 |
Organism: Streptococcus thermophilus, Streptococcus phage vb_sths-va214
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: RNA BINDING PROTEIN |
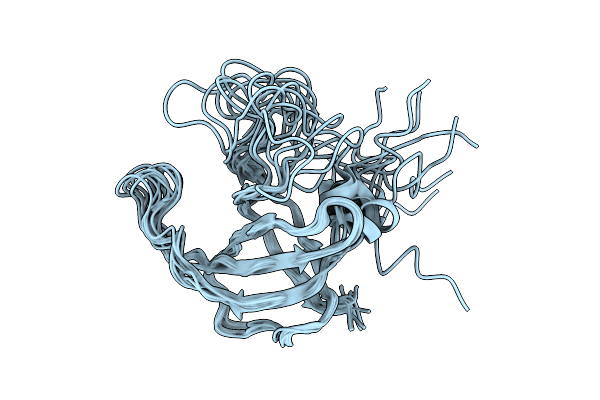 |
Organism: Streptococcus thermophilus
Method: SOLUTION NMR Release Date: 2025-10-22 Classification: NUCLEAR PROTEIN |
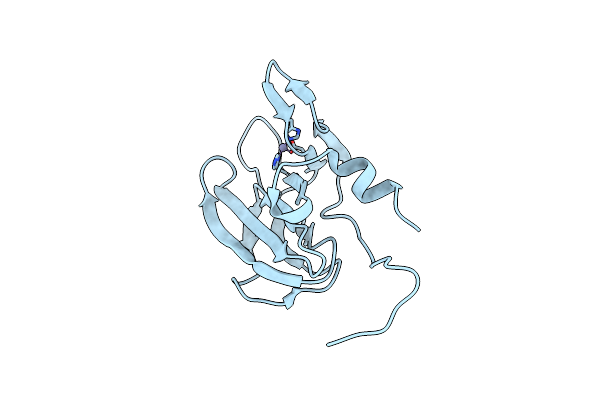 |
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-06-04 Classification: HYDROLASE Ligands: ZN |
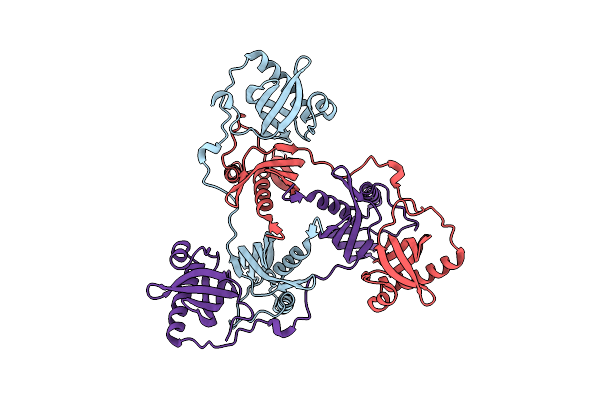 |
Crystal Structure Of A Double Mutant Of Virb8-Like Orfg Central And C-Terminal Domains Of Streptococcus Thermophilus Icest3 (Gram Positive Conjugative Type Iv Secretion System).
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2025-03-12 Classification: TRANSPORT PROTEIN |
 |
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.54 Å Release Date: 2023-10-04 Classification: RNA BINDING PROTEIN |
 |
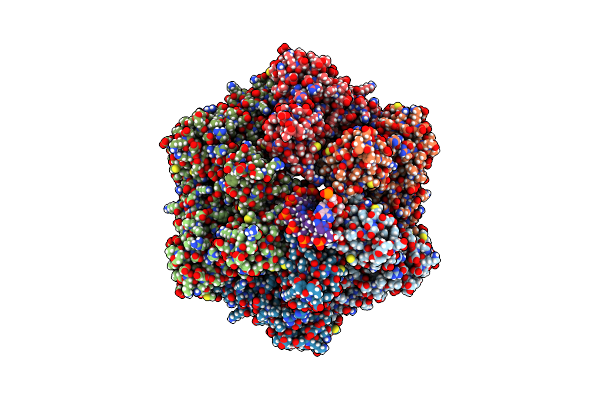
Structure Of Csm6' From Streptococcus Thermophilus In Complex With Cyclic Hexa-Adenylate (Ca6)
Organism: Streptococcus thermophilus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-10-04 Classification: RNA BINDING PROTEIN |
 |
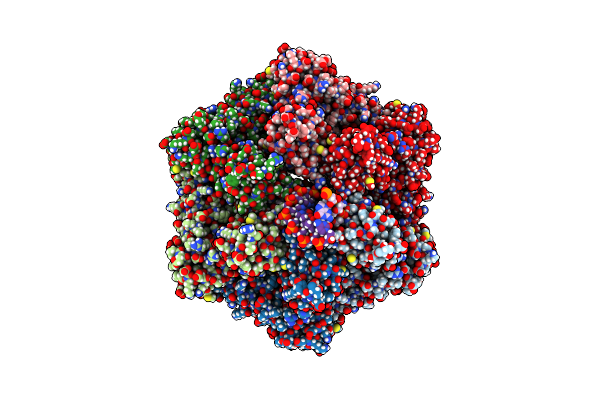
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S1 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
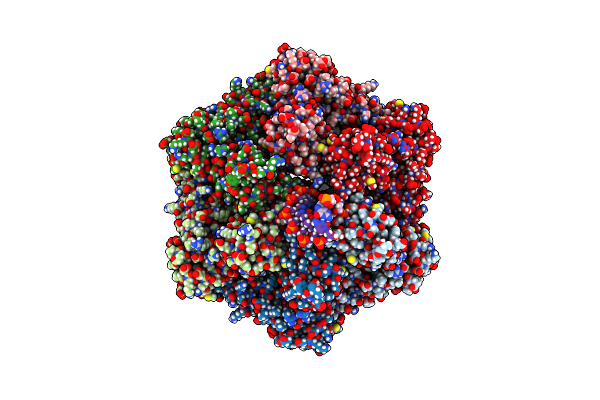
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S2 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
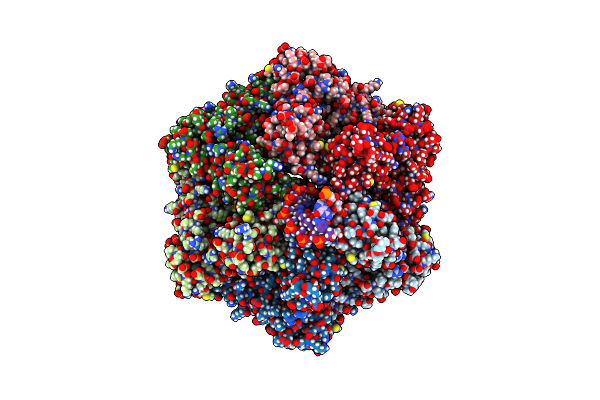
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S3 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, MG, AGS |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S4 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S5 [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: ADP, AGS, MG |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T2 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0-A [T2 Dataset]
Organism: Streptococcus thermophilus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S0+A [T1 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Ruvab Branch Migration Motor Complexed To The Holliday Junction - Ruvb Aaa+ State S1 [T1 Dataset]
Organism: Streptococcus thermophilus, Salmonella typhimurium, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-09-14 Classification: HYDROLASE Ligands: AGS, MG, ADP |
 |
Crystal Structure Of Virb8-Like Orfg Central And C-Terminal Domains Of Streptococcus Thermophilus Icest3 (Gram Positive Conjugative Type Iv Secretion System).
Organism: Streptococcus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2022-09-07 Classification: TRANSPORT PROTEIN Ligands: SO4, GOL |

