Search Count: 183
 |
Crystal Structure Of Lyssavirus Rabies (Ni-Ce Strain) Nucleoprotein In Complex With Phosphoprotein Chaperone
Organism: Rabies virus (strain nishigahara rceh)
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: VIRAL PROTEIN |
 |
Organism: Vibrio xiamenensis
Method: X-RAY DIFFRACTION Release Date: 2025-02-12 Classification: CHAPERONE Ligands: EDO |
 |
Organism: Thermosulfidibacter takaii abi70s6
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-01-22 Classification: TRANSFERASE Ligands: COA, OAA, CIT |
 |
Organism: Thermus phage tsp4
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN Ligands: TMP, MG, TB, CL |
 |
Organism: Rabies virus nishigahara rceh
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN Ligands: SO4 |
 |
Organism: Rabies virus nishigahara rceh
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-04-20 Classification: VIRAL PROTEIN Ligands: EDO, SO4, PO4 |
 |
Crystal Structure Of Rabies Virus (Nishigahara Strain) Phosphoprotein C-Terminal Domain (K214A)
Organism: Rabies virus (strain nishigahara rceh)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-03-17 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Porcine Acbd3 Gold Domain In Complex With 3A Protein Of Aichivirus C
Organism: Sus scrofa, Aichivirus c
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-11-13 Classification: VIRAL PROTEIN Ligands: BGC |
 |
The Crystal Structure Of A Bacterial Aryl Acylamidase Belonging To The Amidase Signature (As) Enzymes Family
Organism: Bacterium csbl00001
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: PO4 |
 |
The Crystal Structure Of A Bacterial Aryl Acylamidase Belonging To The Amidase Signature (As) Enzymes Family
Organism: Bacterium csbl00001
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: CXS, TYL |
 |
Organism: Streptomyces sp. nrrl 2288
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2015-09-23 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
Structure Of Crotonyl-Coa Carboxylase/Reductase Ante V350A In Complex With Nadp
Organism: Streptomyces sp. nrrl 2288
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-09-23 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
2.8 Angstrom Resolution Crystal Structure Of D-Hydantoinase From Bacillus Sp. Ar9 In C2221 Space Group
Organism: Bacillus sp. ar9
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-05-21 Classification: HYDROLASE Ligands: MN |
 |
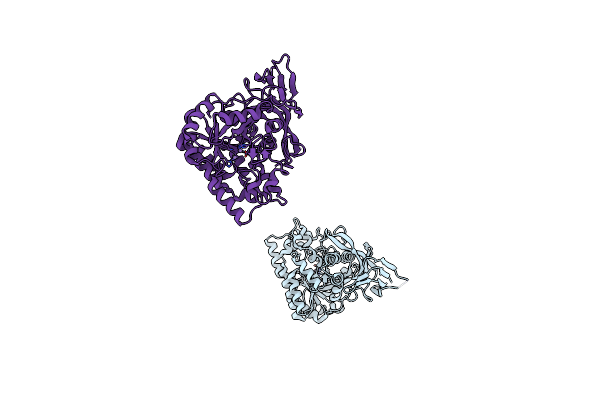
Crystal Structure Of D-Hydantoinase From Bacillus Sp. Ar9 In C2221 Space Group
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-05-07 Classification: HYDROLASE Ligands: MN |
 |
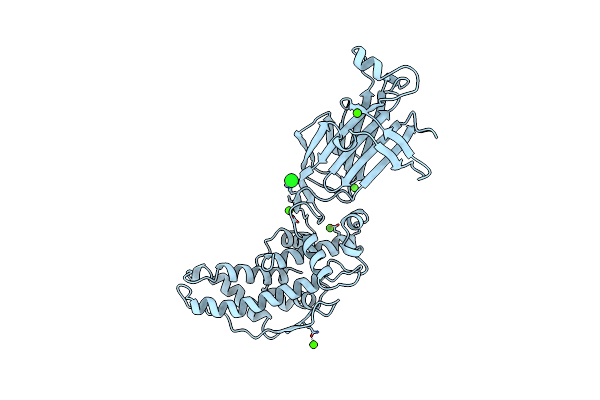
Molecular Structure Of D-Hydantoinase From A Bacillus Sp. Ar9: Evidence For Mercury Inhibition
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-03-01 Classification: HYDROLASE Ligands: MN |
 |
Organism: Bovine rotavirus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2001-04-13 Classification: VIRAL PROTEIN Ligands: ZN, CL, CA |
 |
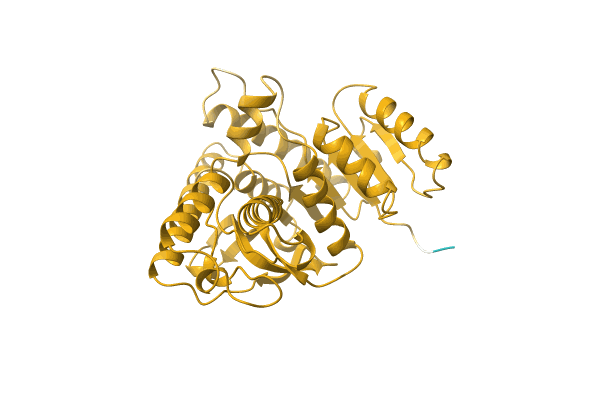 |
Organism: Thermosulfidibacter takaii (strain DSM 17441 / JCM 13301 / NBRC 103674 / ABI70S6)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
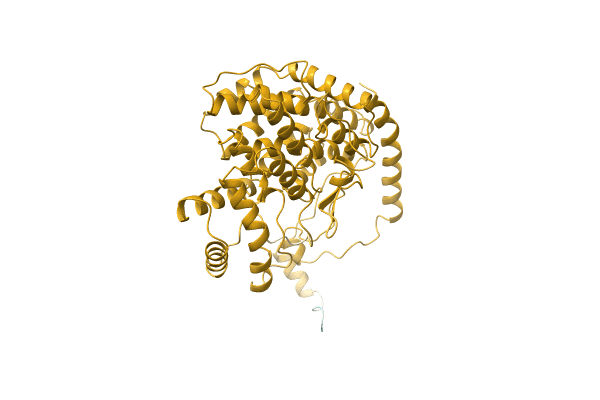 |
Organism: Thermosulfidibacter takaii (strain DSM 17441 / JCM 13301 / NBRC 103674 / ABI70S6)
Method: Alphafold Release Date: Classification: NA Ligands: NA |
 |
Organism: Vibrio genomosp. F6 str. FF-238
Method: Alphafold Release Date: Classification: NA Ligands: NA |

