Search Count: 3,196
 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: HYDROLASE Ligands: PEG, A1EDS |
 |
Organism: Respiratory syncytial virus, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of C36S Mutant Glutathione Peroxidase Of Staphylococcus Aureus.
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: OXIDOREDUCTASE |
 |
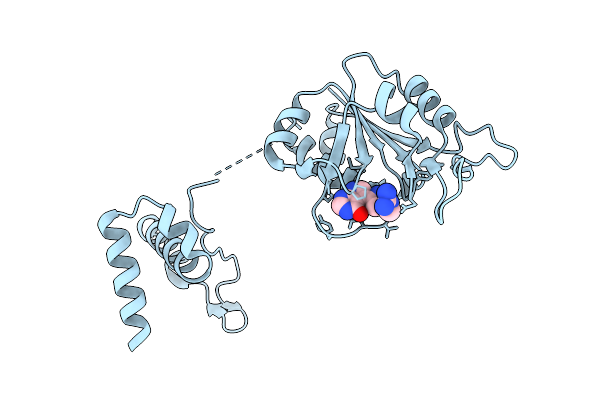
Trna (Guanine-7-)-Methyltransferase (Trmd) From Staphylococcus Aureus In Complex With Sam-Competitive Compound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: EDO, 21X |
 |
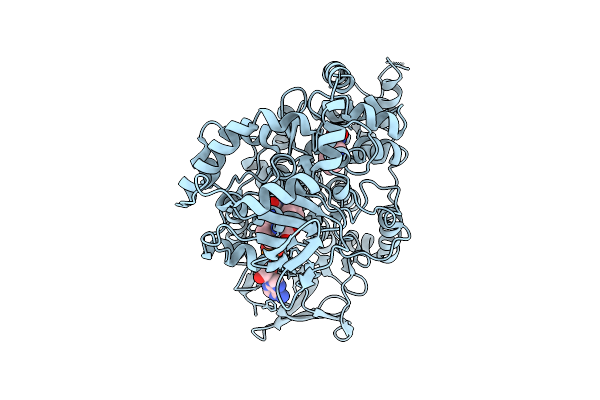
Trna (Guanine-7-)-Methyltransferase (Trmd) From Staphylococcus Aureus In Complex With Sam-Competitive Compound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE Ligands: A1JNG |
 |
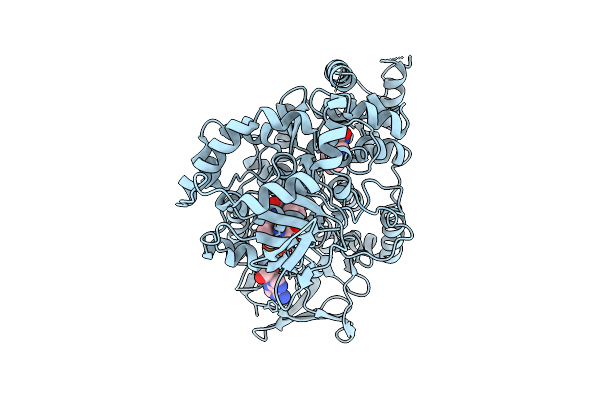
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: FAD, TRP |
 |
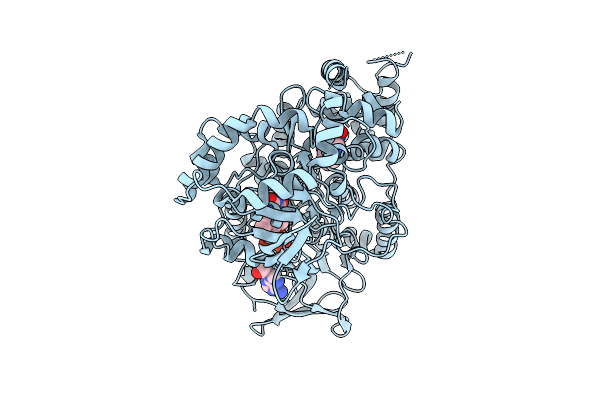
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: FAD, TRP |
 |
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: FAD, TRP |
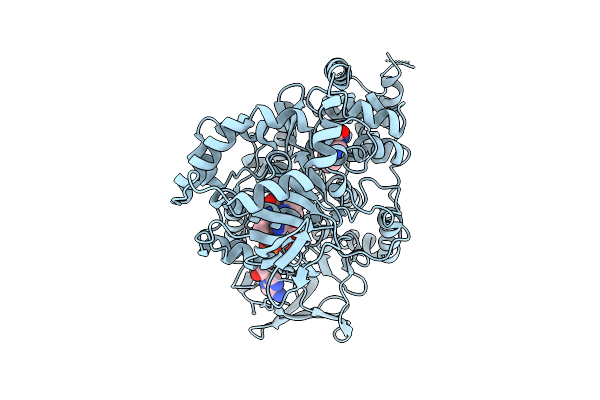 |
Crystal Structure Of Aetf-L183F/V220I/S523A In Complex With Fad And L-Tryptophan
Organism: Aetokthonos hydrillicola thurmond2011
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: FLAVOPROTEIN Ligands: TRP, FAD |
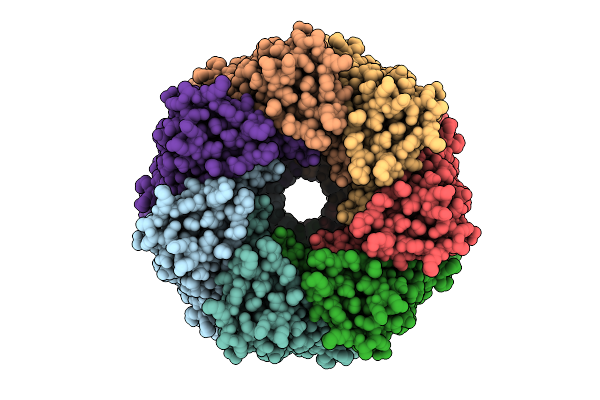 |
Cryo-Em Structure Of Pre-Pore State Iv Of Heptameric Alpha-Hemolysin In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
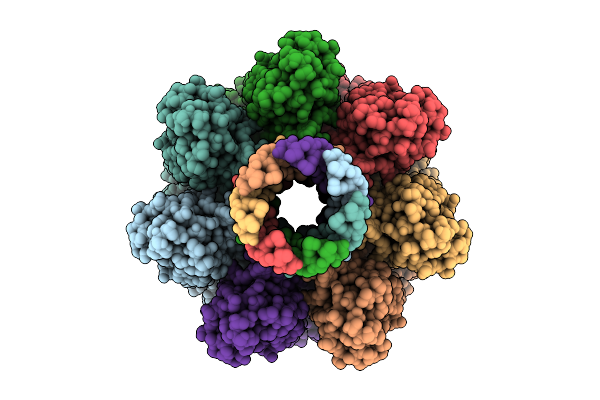 |
Cryo-Em Structure Of Alpha-Hemolysin Heptameric Pore State In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
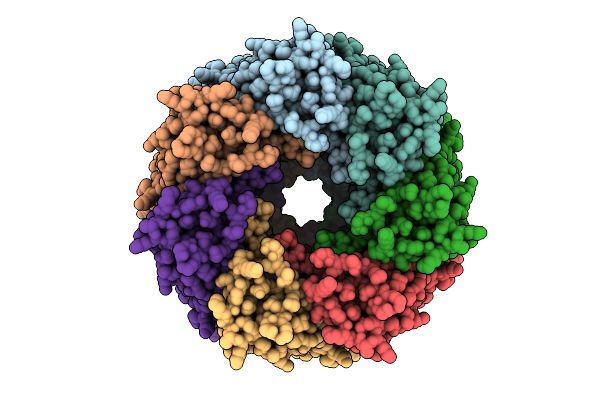 |
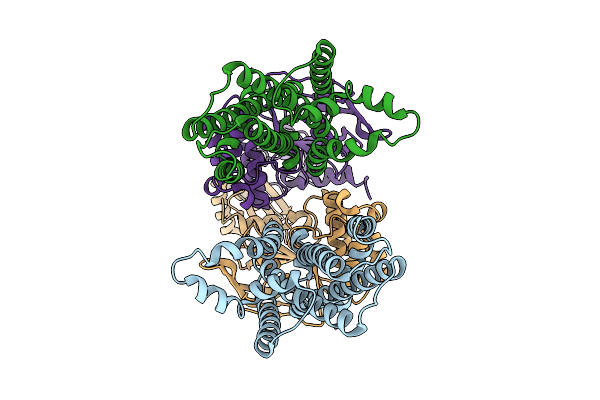
Cryo-Em Structure Of Alpha-Hemolysin Heptameric Pre-Pore State Iii In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN |
 |
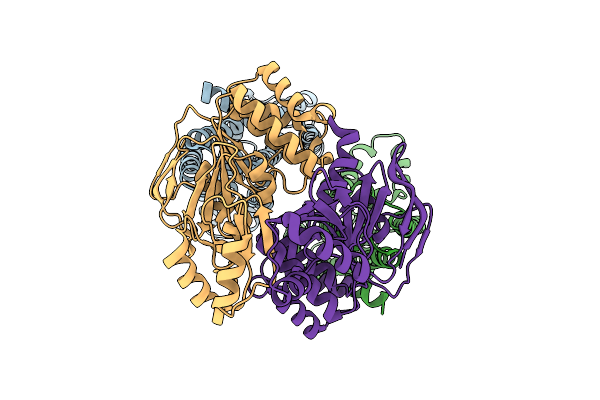
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
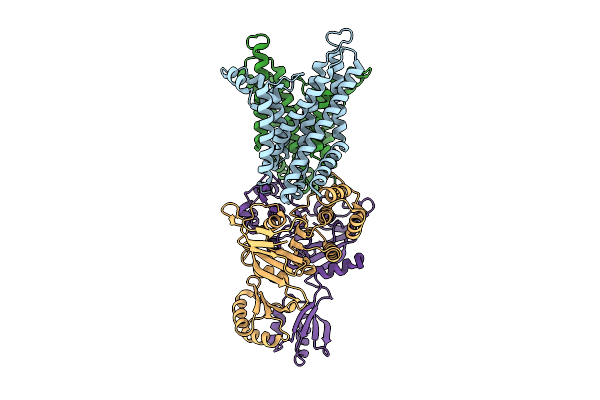 |
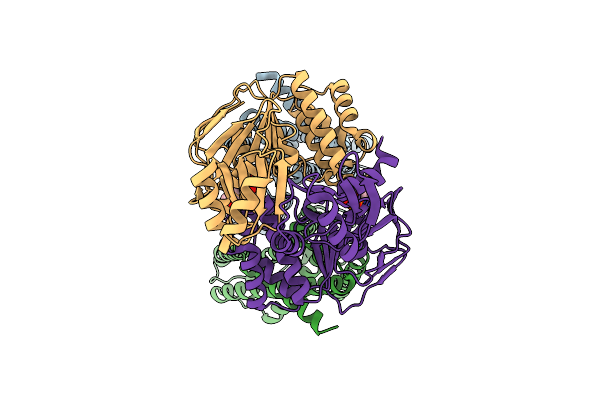
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN |
 |
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: ADP |
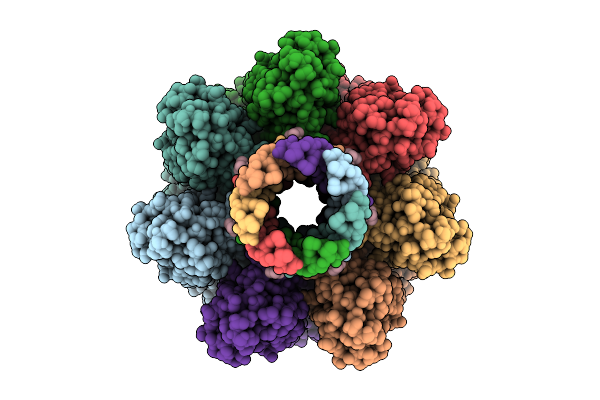 |
Phosphatidylcholine Head Group Bound Alpha-Hemolysin Heptameric Pore Structure In The Presence Of Rbc
Organism: Staphylococcus aureus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: TOXIN Ligands: PC |
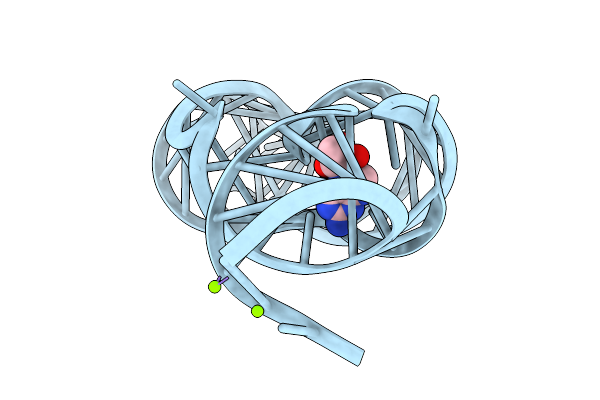 |
Crystal Structure Of Guanine-Ii Riboswitch In Complex With 7,8-Dihydroneopterin
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: NPR, MG |
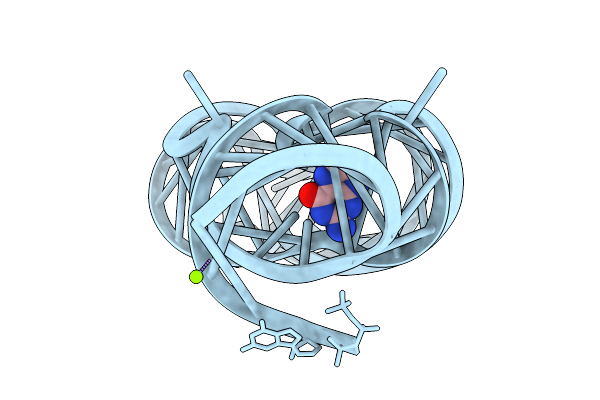 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: 7PD, MG |
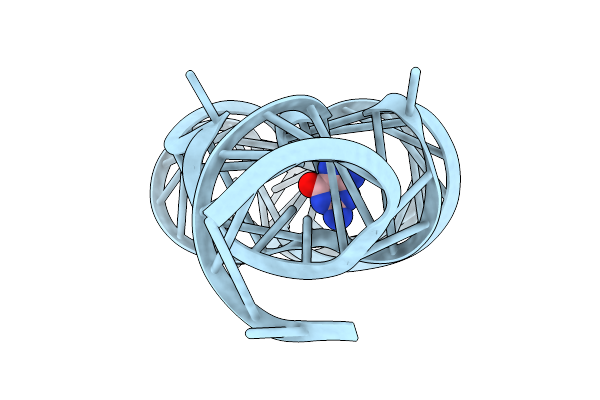 |
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: RNA Ligands: 9QC |

