Search Count: 101
 |
Organism: Acinetobacter terrae, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: PG4 |
 |
Organism: Acinetobacter terrae
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: MEMBRANE PROTEIN Ligands: MG, BTB |
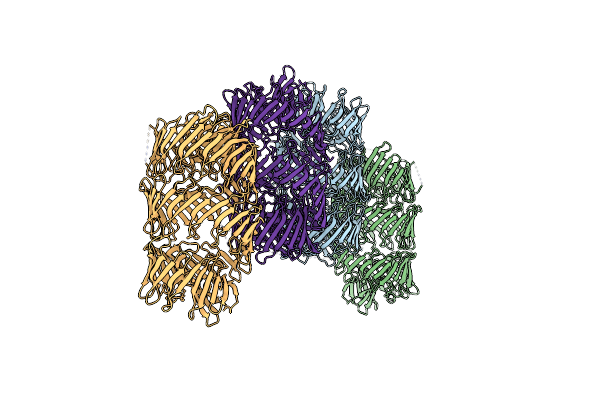 |
T6Ss-Linked Rhs Repeat Protein - Advenella Mimigardefordensis Vgrg-Rhs Core
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:3.36 Å Release Date: 2024-09-25 Classification: CELL INVASION |
 |
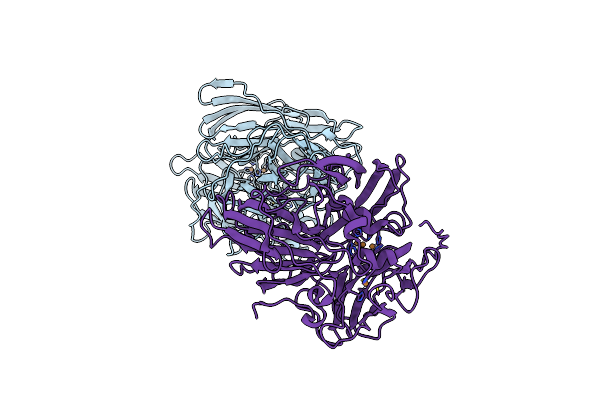
Psychrophilic Laccase (Multicopper Oxidase) From Oenococcus Oeni 229 Without Histag
Organism: Oenococcus oeni
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-08-07 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Psychrophilic Laccase (Multicopper Oxidase) From Oenococcus Oeni 229 With Histag
Organism: Oenococcus oeni
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2024-08-07 Classification: OXIDOREDUCTASE Ligands: CU |
 |
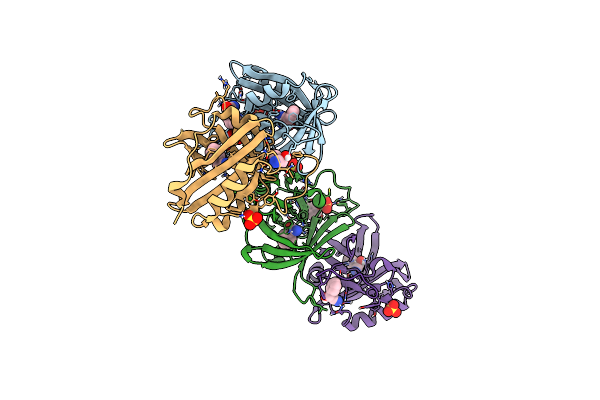
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: HBX, CL, SO4, GOL |
 |
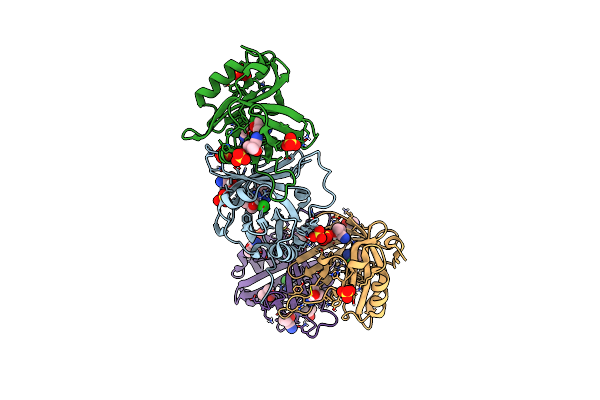
Hydroxynitrile Lyase From The Millipede, Oxidus Gracilis Bound With (R)-(+)-Alpha-Hydroxybenzene-Acetonitrile
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: MXN, SO4 |
 |
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: CNH, CL, SO4 |
 |
Hydroxynitrile Lyase From The Millipede, Oxidus Gracilis Complexed With (R)-2-Chloromandelonitrile
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-24 Classification: LYASE Ligands: IJ5, GOL, SO4 |
 |
Organism: Oxidus gracilis
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2024-01-17 Classification: LYASE Ligands: SO4, CL |
 |
Organism: Arsenophonus endosymbiont of nilaparvata lugens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-06-01 Classification: LIPID TRANSPORT Ligands: OLC |
 |
Organism: Arsenophonus endosymbiont of nilaparvata lugens
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2022-06-01 Classification: LIPID TRANSPORT Ligands: OLC, MPD |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With Xylonic Acid
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-04-14 Classification: TRANSPORT PROTEIN Ligands: TK8 |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With D-Foconate
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-07 Classification: SUGAR BINDING PROTEIN Ligands: TF8 |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With Gluconate
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-07 Classification: SUGAR BINDING PROTEIN Ligands: GCO |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T In Complex With Galactonate
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-04-07 Classification: SUGAR BINDING PROTEIN Ligands: 2Q2 |
 |
Crystal Structure Of The Sugar Acid Binding Protein Dctpam From Advenella Mimigardefordensis Strain Dpn7T
Organism: Advenella mimigardefordensis dpn7
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2021-03-24 Classification: SUGAR BINDING PROTEIN Ligands: KDG |
 |
Cryo-Em Structure Of The Sec Complex From S. Cerevisiae, Wild-Type, Class With Sec62, Conformation 1 (C1)
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: PROTEIN TRANSPORT |
 |
Cryo-Em Structure Of The Sec Complex From S. Cerevisiae, Wild-Type, Class With Sec62, Conformation 2 (C2)
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: PROTEIN TRANSPORT |
 |
Cryo-Em Structure Of The Sec Complex From S. Cerevisiae, Sec61 Pore Mutant, Class With Sec62, Conformation 1 (C1)
Organism: Saccharomyces cerevisiae by4741
Method: ELECTRON MICROSCOPY Release Date: 2021-01-06 Classification: PROTEIN TRANSPORT |

