Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
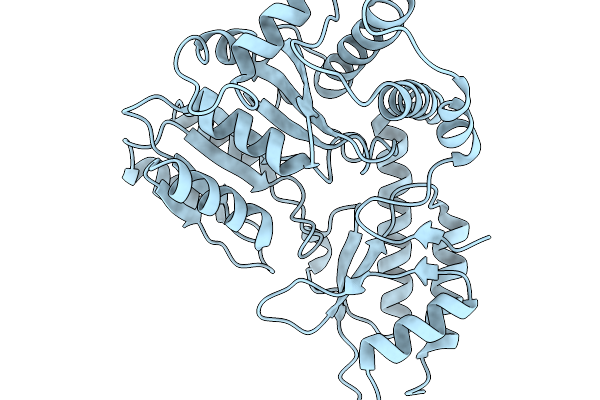 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE |
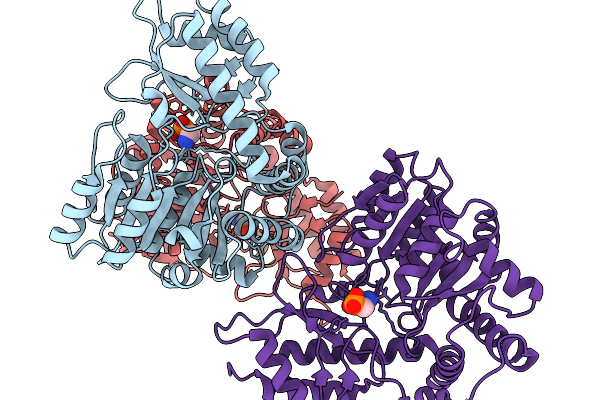 |
Structure Of The Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: P7I |
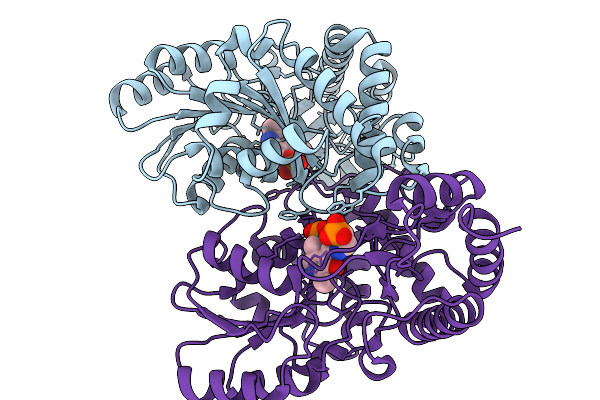 |
Structure Of The K193M Mutant Of Transaminase Phnw From Vibrio Vulnificus In Complex With Plp And Aep
Organism: Vibrio vulnificus nbrc 15645 = atcc 27562
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TRANSFERASE Ligands: PLP, P7I, CL |
 |
Organism: Hbv genotype d3, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
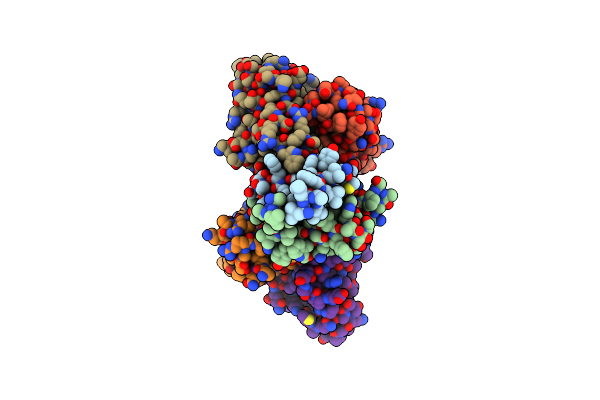 |
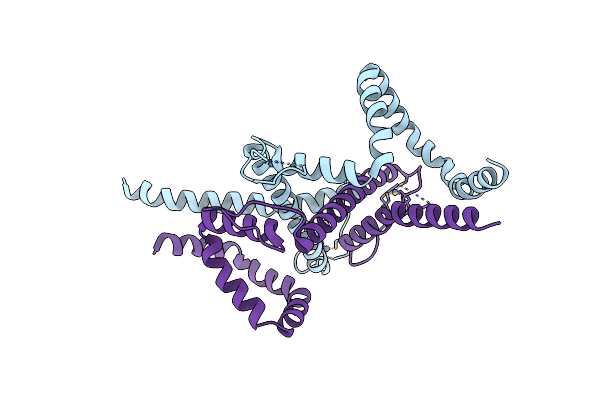
Organism: Homo sapiens, Hbv genotype d3
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: IMMUNE SYSTEM/VIRAL PROTEIN |
 |
Structure Of Hbv Surface Antigen Determined In Recombinant Spherical Subviral Particle
Organism: Hbv genotype d3
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Rhodococcus wratislaviensis nbrc 100605
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: FE, GOL, MG |
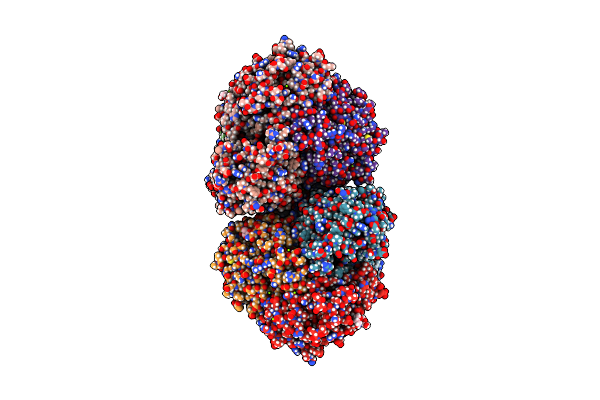 |
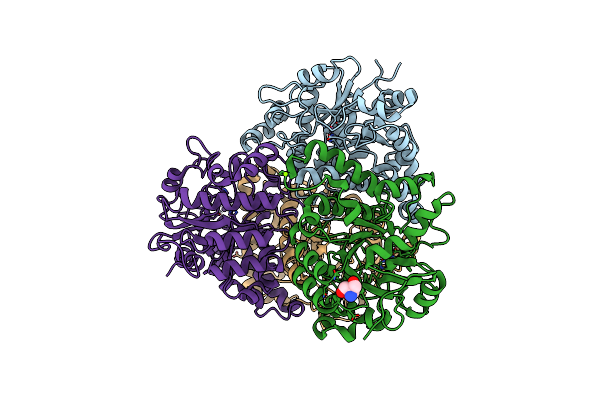
Organism: Rhodococcus wratislaviensis nbrc 100605
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: PG4, FE, MG, TRS, PEG |
 |
Organism: Rhodococcus wratislaviensis nbrc 100605
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: MN, TRS, MG, FE |
 |
Organism: Rhodococcus wratislaviensis nbrc 100605
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2023-04-05 Classification: OXIDOREDUCTASE Ligands: FE, TRS |
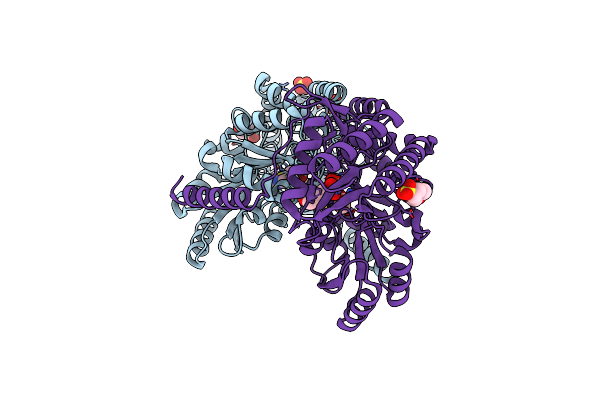 |
Organism: Vibrio proteolyticus nbrc 13287
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-12-29 Classification: TRANSFERASE Ligands: PLP, MES, SO4, GLY |
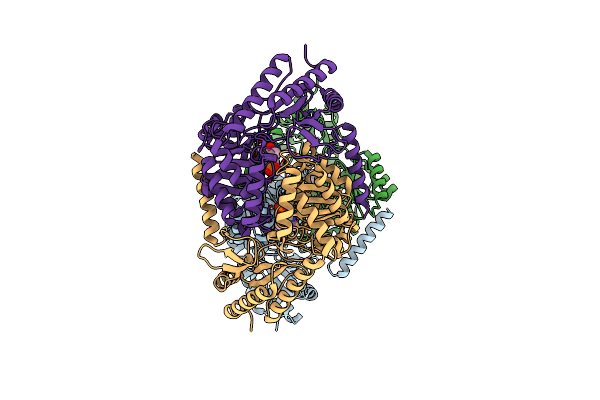 |
Organism: Vibrio proteolyticus nbrc 13287
Method: X-RAY DIFFRACTION Resolution:3.08 Å Release Date: 2021-12-29 Classification: TRANSFERASE Ligands: PLP |
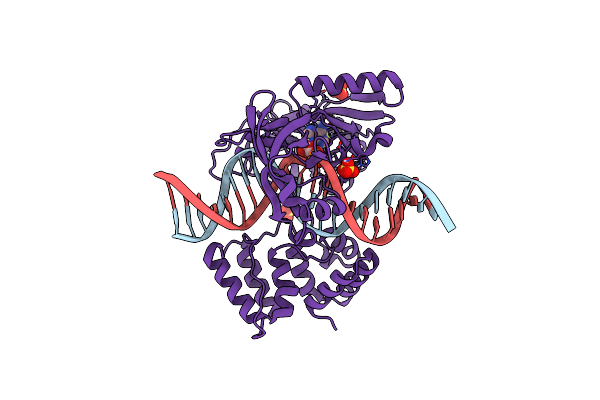 |
Organism: Prochlorococcus marinus, Dna launch vector pde-gfp2
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: AMP, SO4 |
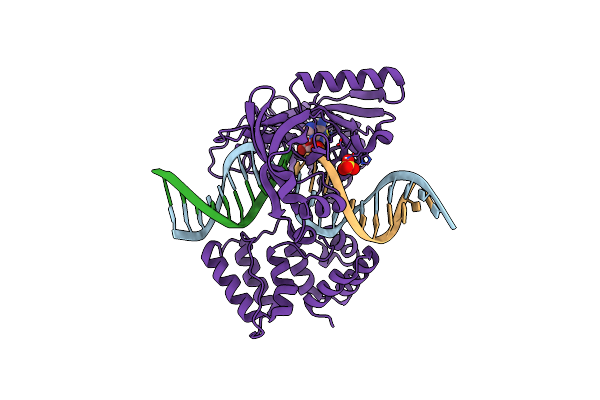 |
Organism: Prochlorococcus marinus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-07-10 Classification: DNA BINDING PROTEIN Ligands: AMP, SO4 |
 |
Organism: Sphingobium herbicidovorans (strain atcc 700291 / dsm 11019 / nbrc 16415 / mh)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE Ligands: CO, AKG |
 |
Organism: Sphingobium herbicidovorans (strain atcc 700291 / dsm 11019 / nbrc 16415 / mh)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-08-15 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Pyranose Dehydrogenase From Agaricus Meleagris, Wildtype
Organism: Leucoagaricus meleagris
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-08-07 Classification: OXIDOREDUCTASE Ligands: FED, NAG, PO4 |
 |
Structure Of The Adpxalf3-Stabilized Transition State Of The Nitrogenase-Like Dark-Operative Protochlorophyllide Oxidoreductase Complex From Prochlorococcus Marinus With Its Substrate Protochlorophyllide A
Organism: Prochlorococcus marinus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-01-30 Classification: OXIDOREDUCTASE Ligands: 1PE, EPE, AF3, ADP, MG, SF4, GOL, PMR, K |
 |
 |