Search Count: 5
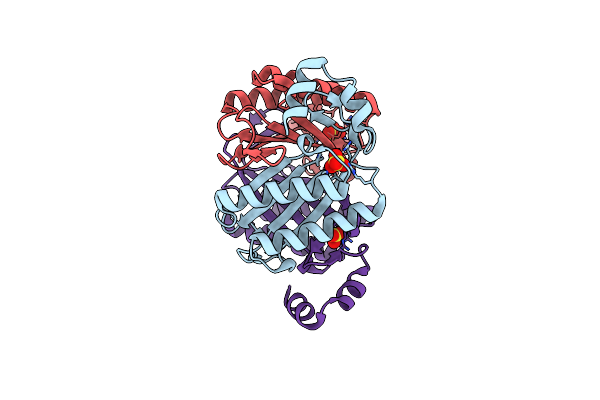 |
Crystal Structure Of N1, A Member Of Cis-3-Chloroacrylic Acid Dehalogenase (Cis-Caad) Family
Organism: Corynebacterium halotolerans yim 70093 = dsm 44683
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-06-02 Classification: HYDROLASE Ligands: SO4 |
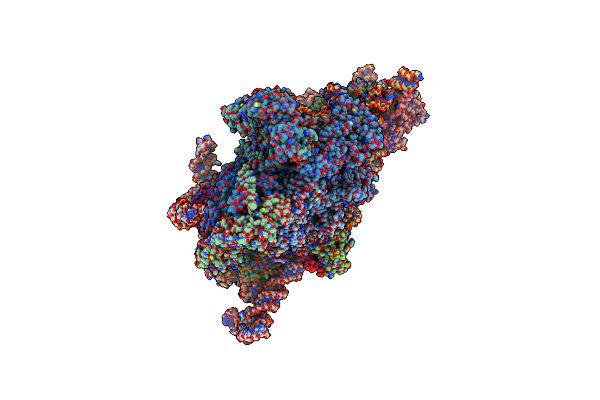 |
Structure Of The Cricket Paralysis Virus 5-Utr Ires (Crpv 5-Utr-Ires) Bound To The Small Ribosomal Subunit In The Open State (Class 1)
Organism: Cricket paralysis virus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Resolution:3.47 Å Release Date: 2020-04-22 Classification: Ribosome/TRANSLATION Ligands: MG, ZN |
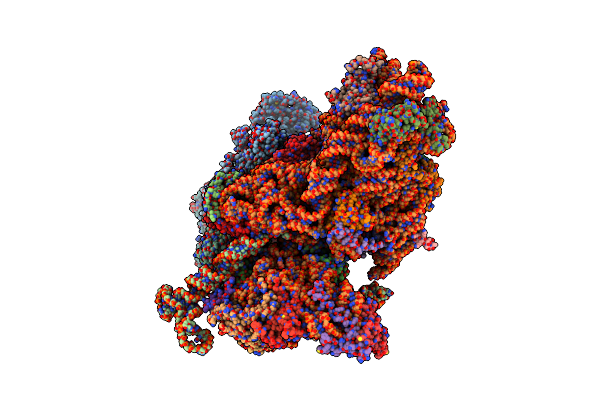 |
Structure Of The Cricket Paralysis Virus 5-Utr Ires (Crpv 5-Utr-Ires) Bound To The Small Ribosomal Subunit In The Closed State (Class 2)
Organism: Cricket paralysis virus, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Resolution:3.36 Å Release Date: 2020-04-22 Classification: RIBOSOME Ligands: MG, ZN |
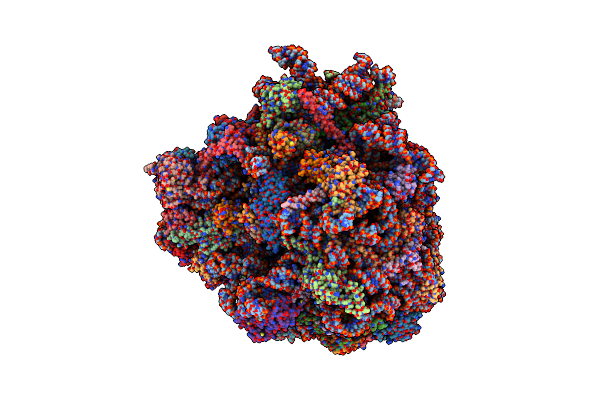 |
Structure Of The Kluyveromyces Lactis 80S Ribosome In Complex With The Cricket Paralysis Virus Ires And Eef2
Organism: Cricket paralysis virus, Saccharomyces cerevisiae p283, Kluyveromyces lactis, Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Release Date: 2016-05-18 Classification: RIBOSOME Ligands: MG, ZN, GCP, 6EM |
 |
Structure Of The Yeast Kluyveromyces Lactis Small Ribosomal Subunit In Complex With The Cricket Paralysis Virus Ires.
Organism: Cricket paralysis virus, Kluyveromyces lactis
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2016-05-18 Classification: RIBOSOME Ligands: MG, ZN |

