Search Count: 7
 |
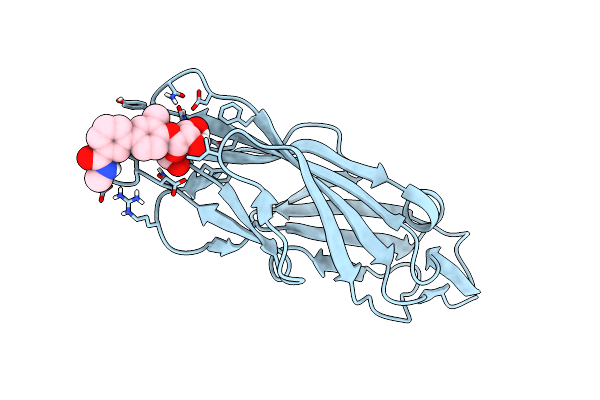
The Cryo-Electron Microscopy Structure Of The Type 1 Chaperone-Usher Pilus Rod
Organism: Escherichia coli j96
Method: ELECTRON MICROSCOPY Release Date: 2017-11-22 Classification: PROTEIN FIBRIL |
 |
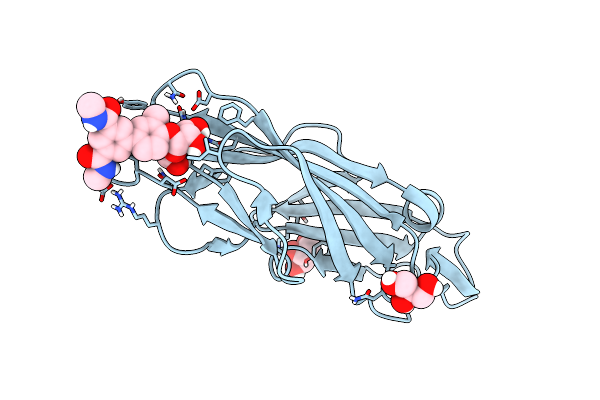
Crystal Structure Of Para-Biphenyl-2-Methyl-3'-Methyl Amide Mannoside Bound To Fimh Lectin Domain
Organism: Escherichia coli j96
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2016-12-07 Classification: SUGAR BINDING PROTEIN Ligands: 5US |
 |
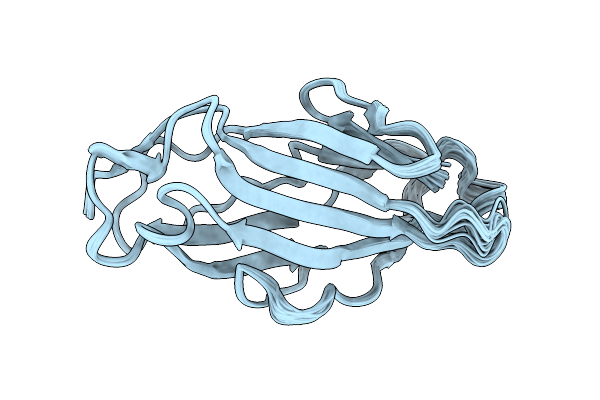
Crystal Structure Of Para-Biphenyl-2-Methyl-3', 5' Di-Methyl Amide Mannoside Bound To Fimh Lectin Domain
Organism: Escherichia coli j96
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2016-05-04 Classification: SUGAR BINDING PROTEIN Ligands: 5U7, GOL |
 |
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2014-02-12 Classification: CELL ADHESION |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2013-07-10 Classification: CELL ADHESION Ligands: BWG, CL |
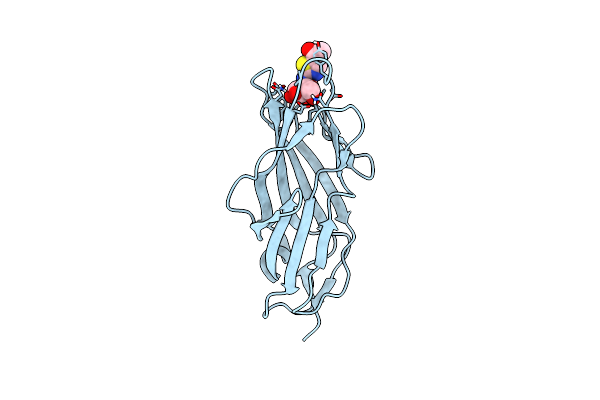 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2013-07-10 Classification: CELL ADHESION Ligands: BWG |
 |
1.7 A Resolution Structure Of The Receptor Binding Domain Of The Fimh Adhesin From Uropathogenic E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2005-02-16 Classification: CELL ADHESION Ligands: DEG, GOL |

