Search Count: 2,202
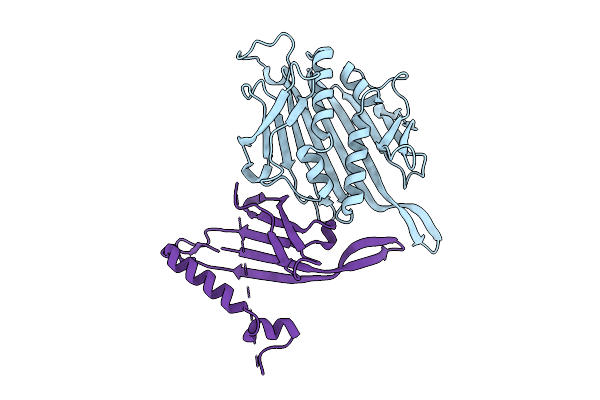 |
Organism: Escherichia phage ms2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRUS LIKE PARTICLE |
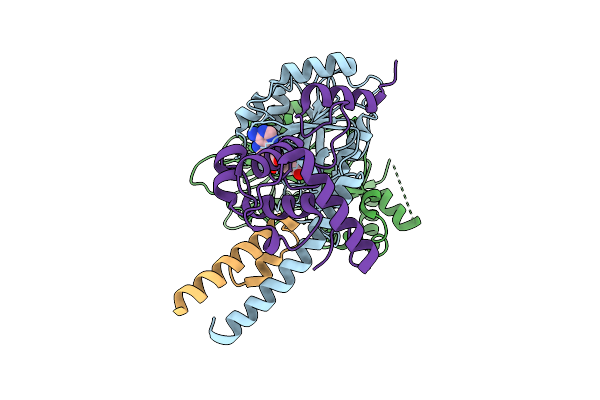 |
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: SAM |
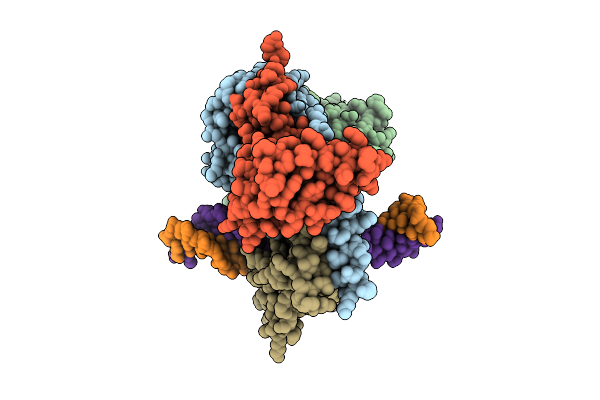 |
Cryo-Em Structure Of Hemimethylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
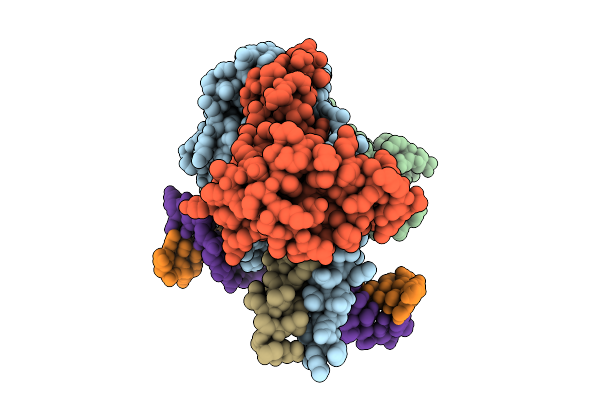 |
Cryo-Em Structure Of Hemi-Methylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C (Mta9-B)
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
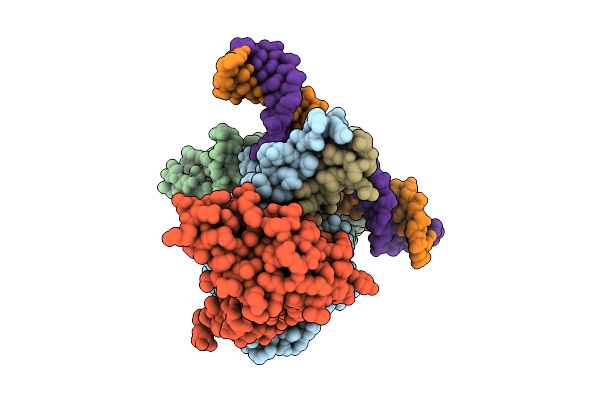 |
Cryo-Em Structure Of Unmethylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C (Mta9-B)
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
 |
Organism: Thalassospira sp.
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: EDO, NPO, SO4, 1PE |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of Estrogen Receptor Alpha From Melanotaenia Fluviatilis Bound To Estradiol And Human Src2 Peptide
Organism: Melanotaenia fluviatilis, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: NUCLEAR PROTEIN Ligands: EST |
 |
Apo Form Of The Estrogen Receptor Alpha Ligand Binding Domain Of Melanotaenia Fluviatilis
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: NUCLEAR PROTEIN Ligands: GOL |
 |
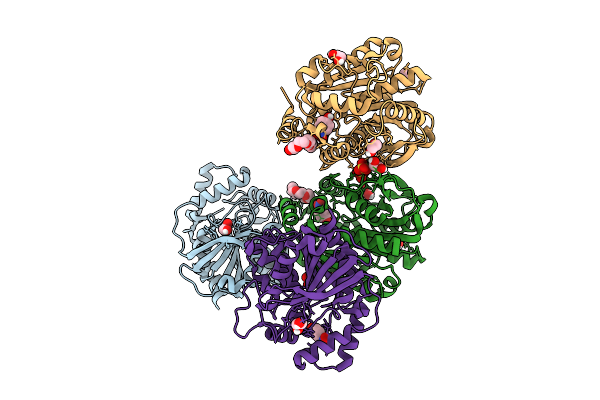
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
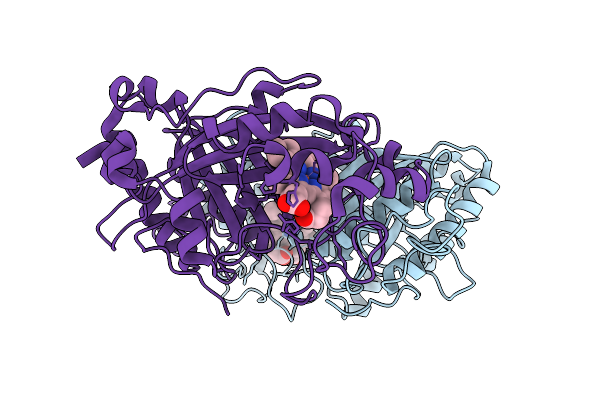 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
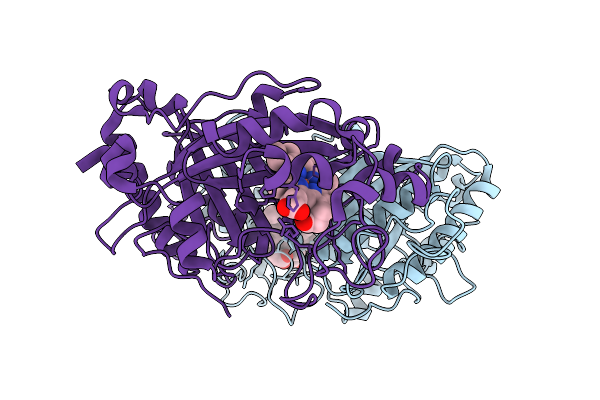 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Streptomyces lividans 1326
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Coxsackievirus b1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: VIRUS LIKE PARTICLE Ligands: MYR |
 |
Crystal Structure Of Cvb3 Replication-Linked Rna Stem-Loop D In Complex With 3C Protein
Organism: Coxsackievirus b3
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/RNA |
 |
Crystal Structure Of Cvb3 Replication-Linked Rna In Complex With 3C Protein
Organism: Coxsackievirus b3
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: VIRAL PROTEIN/RNA Ligands: PO4 |

