Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
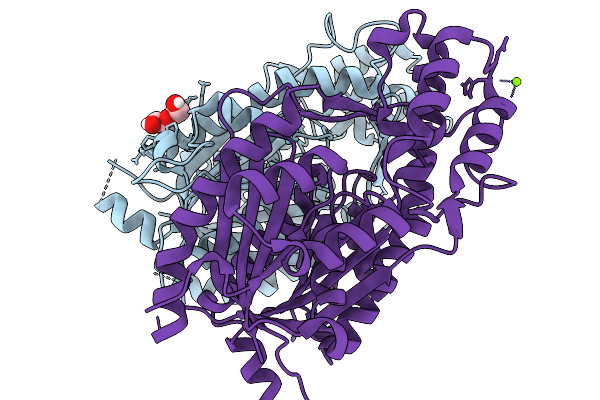 |
A Canonical Homodimer Of Type Iii Polyketide Synthase From Aspergillus Thesauricus Ibt 34227
Organism: Aspergillus thesauricus
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: PEG, MG |
 |
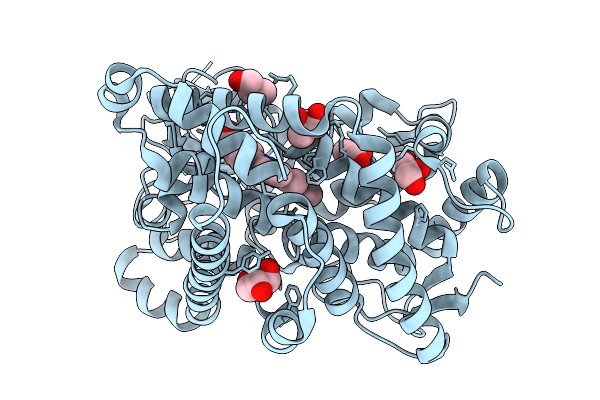
Crystal Structure Of Bmp7 In Complex With 2,4-Dibromophenol Generated By Substrate Soaking
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: GOL, CL, HEM, Y8I |
 |
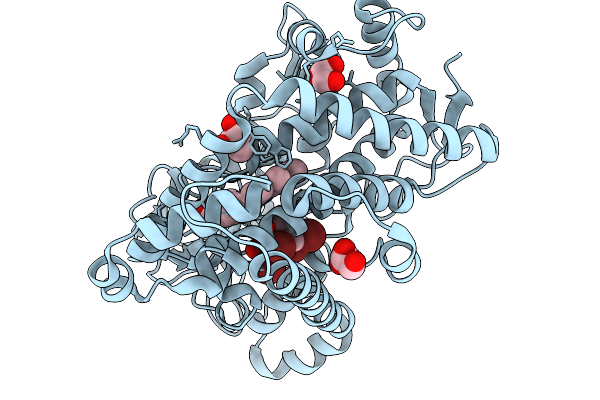
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: HEM, GOL |
 |
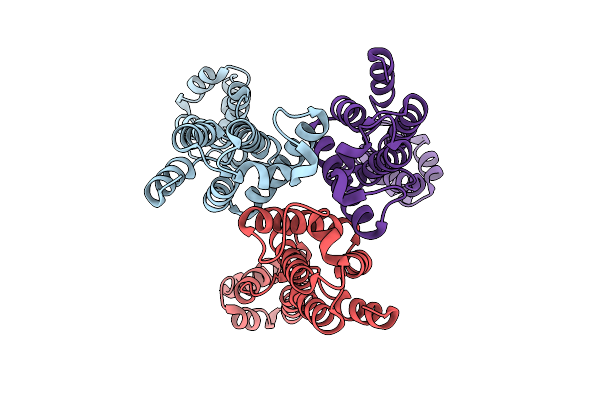
Organism: Marinomonas mediterranea
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: OXIDOREDUCTASE Ligands: GOL, CL, HEM, Y8I |
 |
Organism: Human t-cell leukemia virus type i
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN |
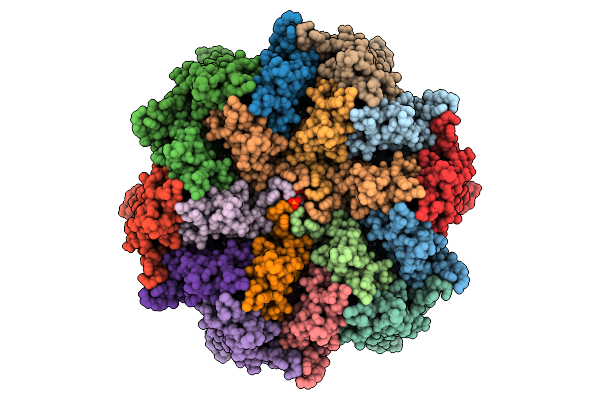 |
High-Resolution Analysis Of The Human T-Cell Leukemia Virus Capsid Protein Reveals Insights Into Immature Particle Morphology
Organism: Human t-cell leukemia virus type i
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRUS LIKE PARTICLE Ligands: IHP |
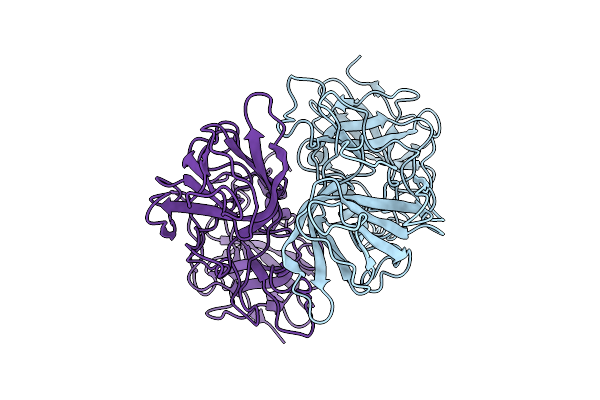 |
Organism: Rabbit hemorrhagic disease virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: VIRAL PROTEIN |
 |
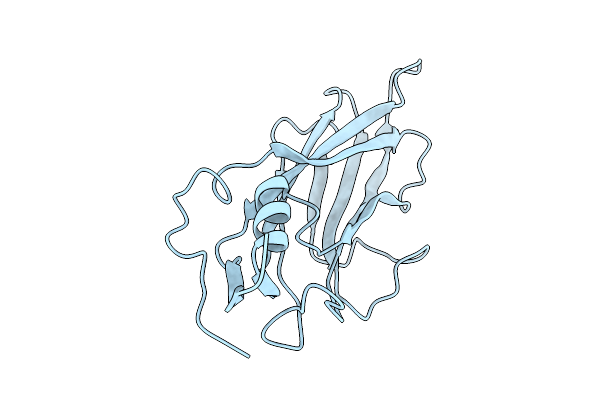
Organism: Rabbit hemorrhagic disease virus
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: VIRAL PROTEIN Ligands: EDO, NHE, TRS, PEG |
 |
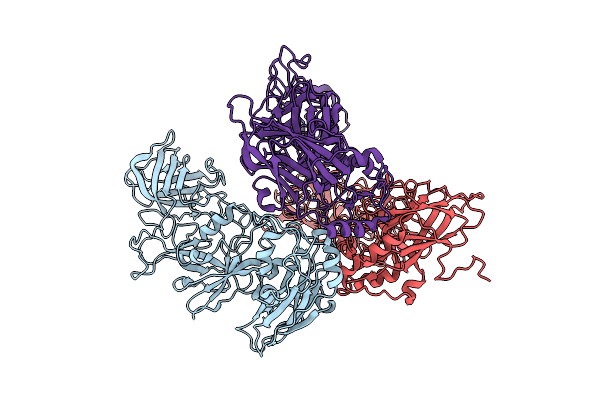
Organism: Rabbit hemorrhagic disease virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS LIKE PARTICLE |
 |
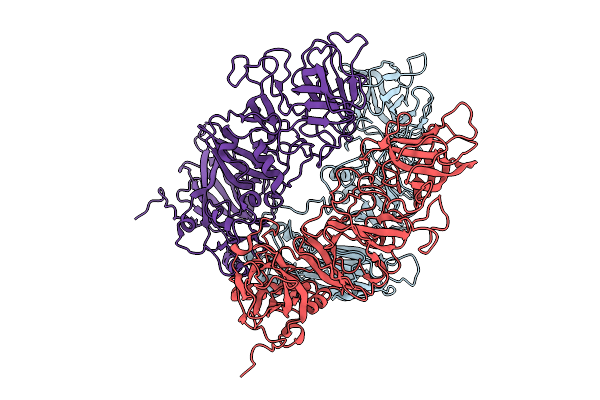
Organism: Rabbit hemorrhagic disease virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRAL PROTEIN |
 |
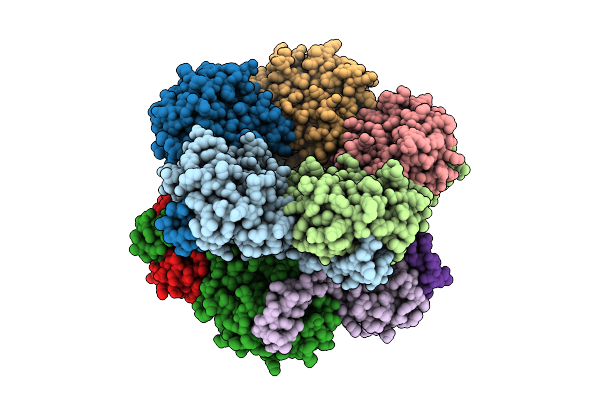
Organism: Rabbit hemorrhagic disease virus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS |
 |
Selective Production Of Versatile L-Glyceraldehyde From C1 And/Or C2 Aldehydes
Organism: Gilliamella apicola
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Norwalk virus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: VIRAL PROTEIN Ligands: AGS, MG |
 |
Organism: Caldanaerobacter subterraneus subsp. tengcongensis
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: ISOMERASE Ligands: B12, 5AD |
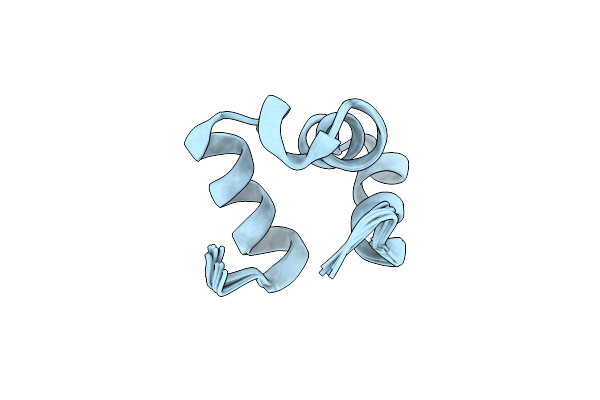 |
Organism: Turnip crinkle virus
Method: SOLUTION NMR Release Date: 2025-08-20 Classification: RNA BINDING PROTEIN |
 |
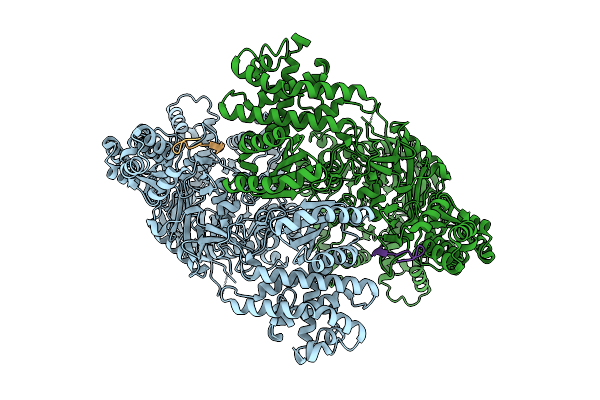
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley In The Apo State
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley, In Complex With Its Substrate
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Acetivibrio thermocellus atcc 27405
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Acetivibrio thermocellus dsm 1313
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: SUGAR BINDING PROTEIN |
 |
Crystal Structure Of The Complex Formed Between The Radical Sam Protein Chlb And The Leader Region Of Its Precursor Substrate Chla
Organism: Fischerella
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: SF4, FMT, SAH, TRS, ZN |