Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
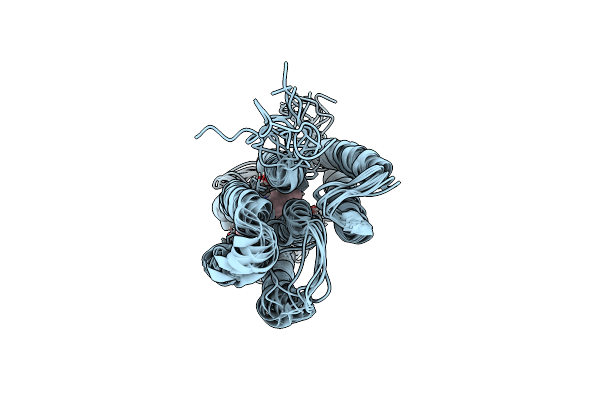 |
Anabaena Sensory Rhodopsin Structure Determination From Paramagnetic Relaxation Enhancement And Nmr Restraints
Organism: Nostoc sp.
Method: SOLID-STATE NMR Release Date: 2025-04-16 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Crystal Structure Of Mutant Nonpro1 Tautomerase Superfamily Member Nj7-V1P In Complex With 3-Bromopropiolate Inhibitor
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-03-05 Classification: ISOMERASE/INHIBITOR Ligands: A1AWY |
 |
Organism: Moloney murine leukemia virus, Streptococcus pyogenes serotype m1, Streptococcus pyogenes
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
Organism: Streptococcus pyogenes serotype m1, Moloney murine leukemia virus, Streptococcus pyogenes
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: RNA BINDING PROTEIN |
 |
Organism: Streptococcus pyogenes serotype m1, Streptococcus pyogenes, Moloney murine leukemia virus
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: RNA BINDING PROTEIN/RNA/DNA |
 |
Organism: Streptococcus pyogenes, Moloney murine leukemia virus, Streptococcus pyogenes serotype m1
Method: ELECTRON MICROSCOPY Release Date: 2024-06-05 Classification: RNA BINDING PROTEIN/DNA/RNA |
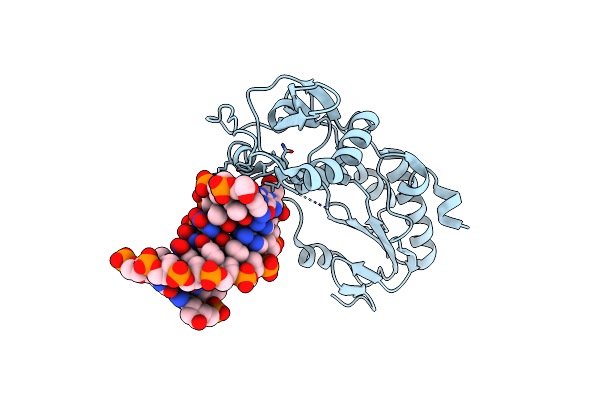 |
Crystal Structure Of Alien Dna Ctszzpbsbszppbag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-17 Classification: DNA BINDING PROTEIN/DNA |
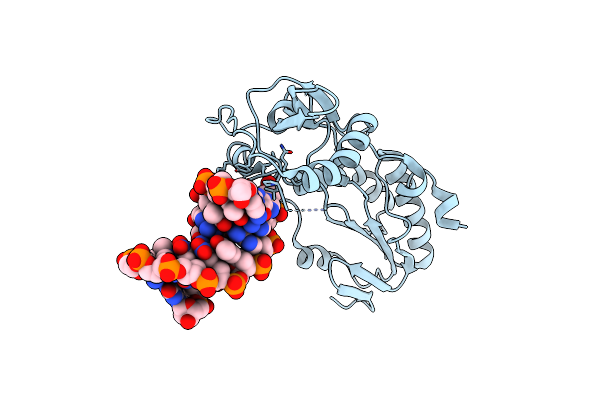 |
Crystal Structure Of B-Form Alien Dna 5'-Cttbppbbsszzsaag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-10 Classification: DNA BINDING PROTEIN/DNA |
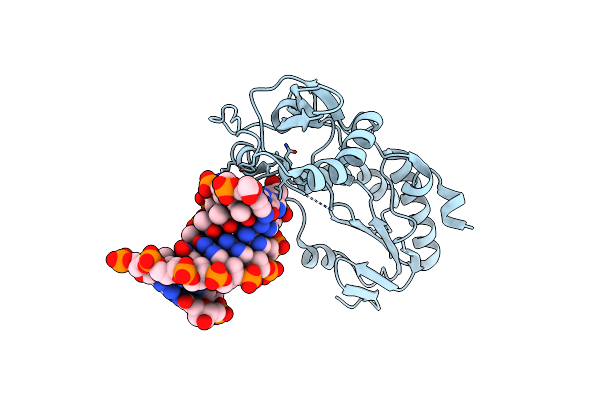 |
Crystal Structure Of B-Form Alien Dna 5'-Cttsspbzpszbbaag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-05-10 Classification: DNA BINDING PROTEIN/DNA |
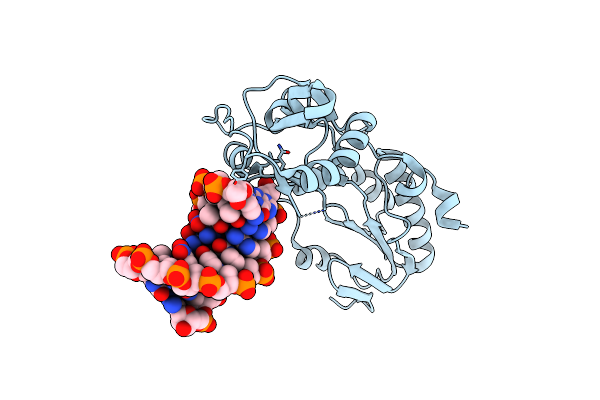 |
Crystal Structure Of B-Form Alien Dna 5'-Cttzzpbsbszppaag In A Host-Guest Complex With The N-Terminal Fragment Of Moloney Murine Leukemia Virus Reverse Transcriptase
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-05-10 Classification: DNA BINDING PROTEIN/DNA |
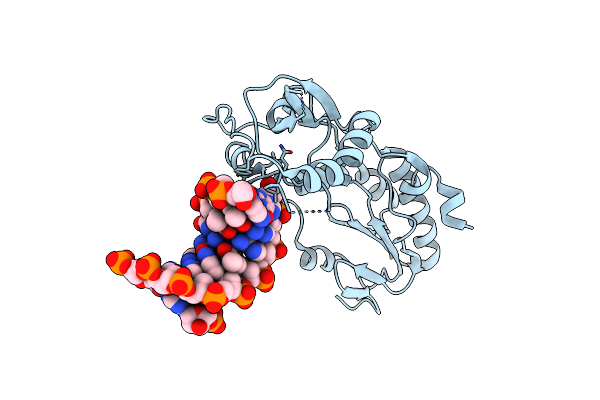 |
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-01-11 Classification: DNA BINDING PROTEIN/DNA |
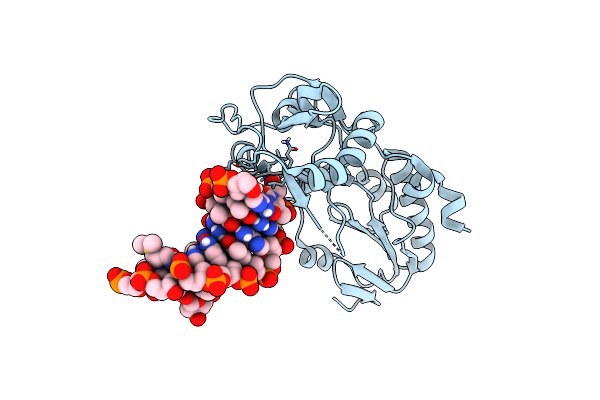 |
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2023-01-11 Classification: DNA BINDING PROTEIN/DNA Ligands: BLM |
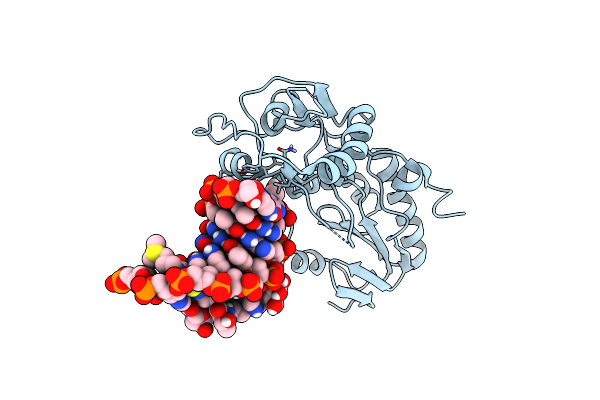 |
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2023-01-11 Classification: DNA BINDING PROTEIN/DNA Ligands: 3CO, BLM |
 |
Solution Nmr Structure And Dynamics Of Human Brd3 Et In Complex With Mlv In Ctd
Organism: Moloney murine leukemia virus, Homo sapiens
Method: SOLUTION NMR Release Date: 2021-06-23 Classification: SIGNALING PROTEIN |
 |
Hydrogen Bonding Complementary, Not Size Complementarity Is Key In The Formation Of The Double Helix
Organism: Moloney murine leukemia virus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-09-19 Classification: DNA BINDING PROTEIN/DNA |
 |
Hydrogen Bonding Complementary, Not Size Complementarity Is Key In The Formation Of The Double Helix
Organism: Moloney murine leukemia virus, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2018-09-19 Classification: DNA BINDING PROTEIN/DNA |
 |
Hydrogen Bonding Complementary, Not Size Complementarity Is Key In The Formation Of The Double Helix
Organism: Moloney murine leukemia virus, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-19 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Nostoc sp.
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-07-11 Classification: SIGNALING PROTEIN Ligands: HEM |
 |
Organism: Squalimorphii
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2017-02-22 Classification: HYDROLASE ACTIVATOR |
 |
Crystal Structure Of The N-Terminal Domain Of Moloney Murine Leukemia Virus Integrase, Northeast Structural Genomics Consortium Target Or3
Organism: Moloney murine leukemia virus
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2010-07-14 Classification: VIRAL PROTEIN Ligands: ZN, ACT |