Search Count: 23
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 1
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4 |
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 2
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 3
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP |
 |
Cryo-Em Reveals Transition States Of The Acinetobacter Baumannii F1-Atpase Rotary Subunits Gamma And Epsilon And Novel Compound Targets - Conformation 4
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4 |
 |
Organism: Acinetobacter baumannii ab5075
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: HYDROLASE Ligands: ATP, MG, ADP, PO4 |
 |
Lymphocytic Choriomeningitis Virus Glycoprotein In Complex With Neutralizing Antibody M28
Organism: Lymphocytic choriomeningitis virus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Lymphocytic choriomeningitis virus
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Clostridium phage phicpv4
Method: ELECTRON MICROSCOPY Release Date: 2022-10-12 Classification: VIRAL PROTEIN |
 |
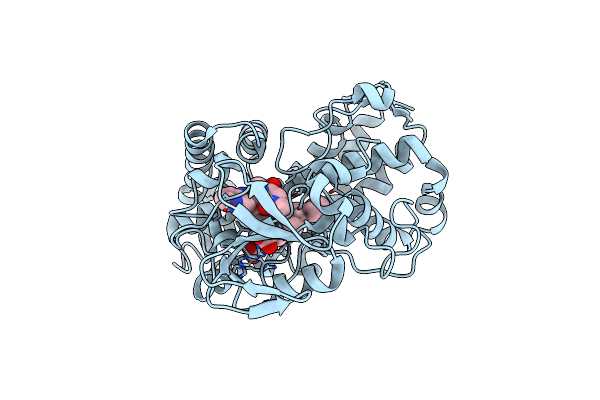
Crystal Structure Of The Murine Class I Major Histocompatibility Complex H-2Db In Complex With Lcmv-Derived Gp33 Peptide With D-Aminoacid
Organism: Mus musculus, Lymphocytic choriomeningitis mammarenavirus
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2022-03-23 Classification: IMMUNE SYSTEM Ligands: SO4, CL |
 |
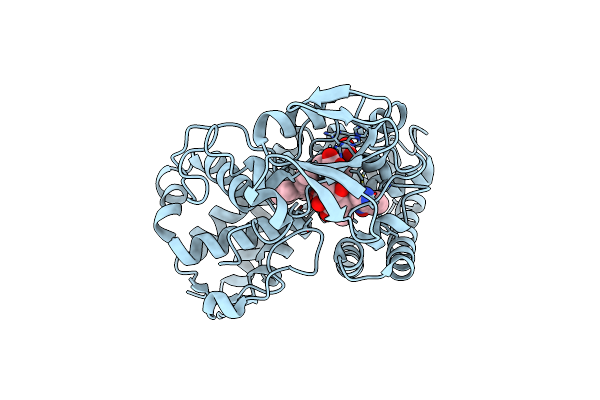
Cytochrome P450 Txtc Employs Substrate Conformational Switching For Sequential Aliphatic And Aromatic Thaxtomin Hydroxylation
Organism: Streptomyces acidiscabies 84-104
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: HEM, C8H |
 |
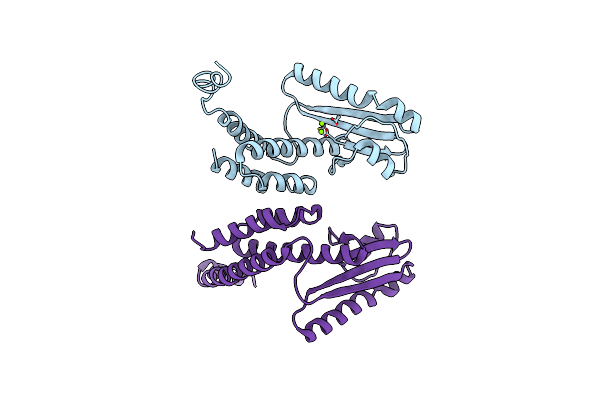
Cytochrome P450 Txtc Employs Substrate Conformational Switching For Sequential Aliphatic And Aromatic Thaxtomin Hydroxylation
Organism: Streptomyces acidiscabies 84-104
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-12-12 Classification: OXIDOREDUCTASE Ligands: HEM, C8B, OXY |
 |
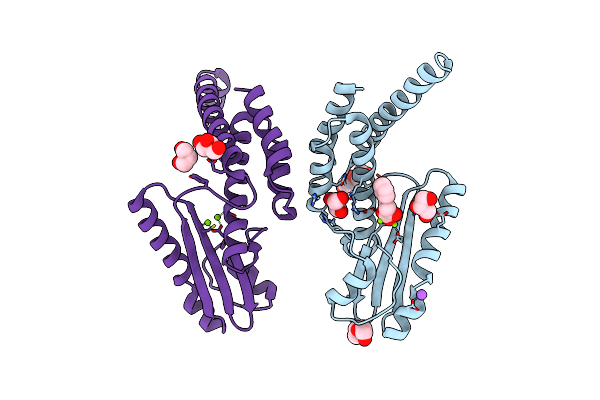
Crystal Structure Of Lymphocytic Choriomeningitis Mammarenavirus Endonuclease Complexed With Catalytic Ions
Organism: Lymphocytic choriomeningitis mammarenavirus
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2017-09-13 Classification: TRANSFERASE Ligands: MG |
 |
Crystal Structure Of Lymphocytic Choriomeningitis Mammarenavirus Endonuclease Complexed With Dpba
Organism: Lymphocytic choriomeningitis mammarenavirus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2017-09-13 Classification: TRANSFERASE Ligands: MG, GOL, XI7, CL, PEG, NA |
 |
Crystal Structure Of Lymphocytic Choriomeningitis Mammarenavirus Endonuclease Mutant D118A
Organism: Lymphocytic choriomeningitis mammarenavirus
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2017-09-13 Classification: TRANSFERASE Ligands: CL, NA, GOL |
 |
Crystal Structure Of Lymphocytic Choriomeningitis Mammarenavirus Endonuclease Bound To Compound L742001
Organism: Lymphocytic choriomeningitis mammarenavirus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2017-09-13 Classification: TRANSFERASE Ligands: MN, 0N8, GOL, CL, NA |
 |
Organism: Lymphocytic choriomeningitis mammarenavirus
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2016-04-20 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of A Putative Phosphomethylpyrimidine Kinase From Acinetobacter Baumannii In Non-Covalent Complex With Pyridoxal Phosphate
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2016-03-02 Classification: TRANSFERASE Ligands: PLP |
 |
Structure Of The Nucleotide Binding Domain Of An Abc Transporter Msba From Acinetobacter Baumannii
Organism: Acinetobacter baumannii ab5075
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-03-02 Classification: TRANSPORT PROTEIN Ligands: EDO |
 |
X-Ray Crystal Structure Of Phosphoglycerate Kinase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-06-17 Classification: TRANSFERASE |
 |
Organism: Acinetobacter baumannii ab5075
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-06-03 Classification: LYASE Ligands: MG, EDO, CL |

