Search Count: 207
 |
Organism: Homo sapiens, Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: ACT, NA |
 |
Organism: Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: IPA |
 |
Organism: Homo sapiens, Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: CIT, PEG |
 |
Crystal Structure Of Bacteroides Ovatus Kdui1 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: ISOMERASE Ligands: GOL |
 |
Crystal Structure Of Bacteroides Ovatus Kdui2 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: ISOMERASE |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Complexed With Nad+ Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Structure Of Uncleaved Influenza A/Victoria/2570/2019 Haemagglutinin. The 2021 Influenza A(H1N1)Pdm09 Egg-Derived Vaccine Candidate.
Organism: H1n1 subtype
Method: ELECTRON MICROSCOPY Release Date: 2025-10-08 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Crystal Structure Of Co(Ii)-Bound Polysaccharide Deacetylase From Bacteroides Ovatus
Organism: Bacteroides ovatus (strain atcc 8483 / dsm 1896 / jcm 5824 / bcrc 10623 / ccug 4943 / nctc 11153)
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: GOL, CO |
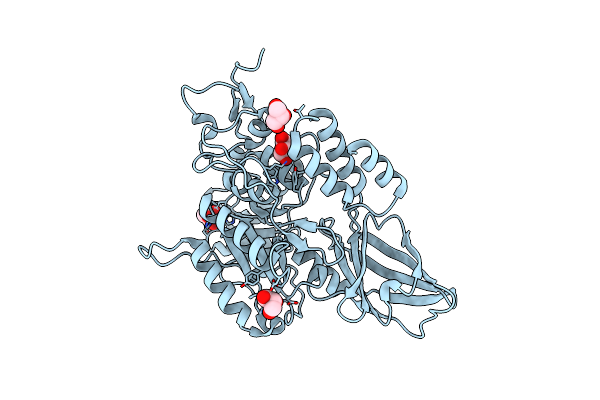 |
Crystal Structure Of Ni(Ii)-Bound Polysaccharide Deacetylase From Bacteroides Ovatus
Organism: Bacteroides ovatus (strain atcc 8483 / dsm 1896 / jcm 5824 / bcrc 10623 / ccug 4943 / nctc 11153)
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: GOL, NI |
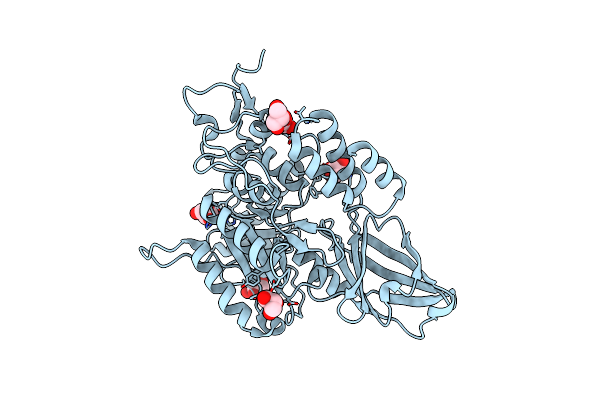 |
Crystal Structure Of Cu(Ii)-Bound Polysaccharide Deacetylase From Bacteroides Ovatus
Organism: Bacteroides ovatus (strain atcc 8483 / dsm 1896 / jcm 5824 / bcrc 10623 / ccug 4943 / nctc 11153)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: GOL, CU |
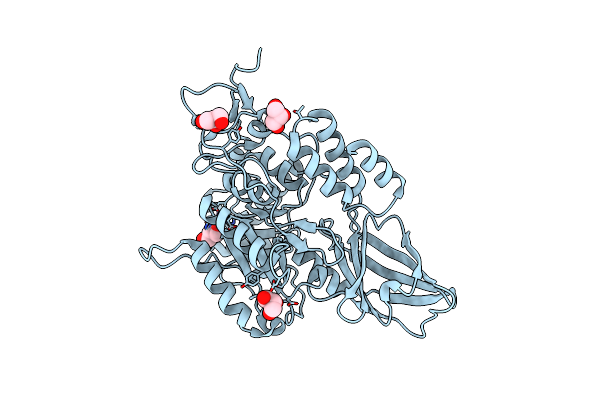 |
Crystal Structure Of Zn(Ii)-Bound Polysaccharide Deacetylase From Bacteroides Ovatus
Organism: Bacteroides ovatus (strain atcc 8483 / dsm 1896 / jcm 5824 / bcrc 10623 / ccug 4943 / nctc 11153)
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: GOL, ZN |
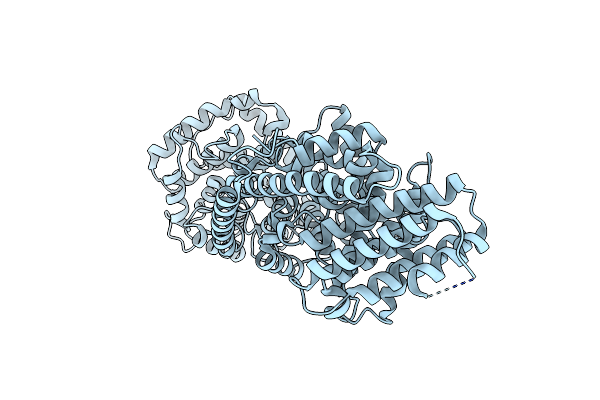 |
Organism: Grindelia hirsutula
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-11-06 Classification: PLANT PROTEIN |
 |
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-10-30 Classification: HYDROLASE Ligands: EDO, PEG, FMT, PGE, CA, NA |
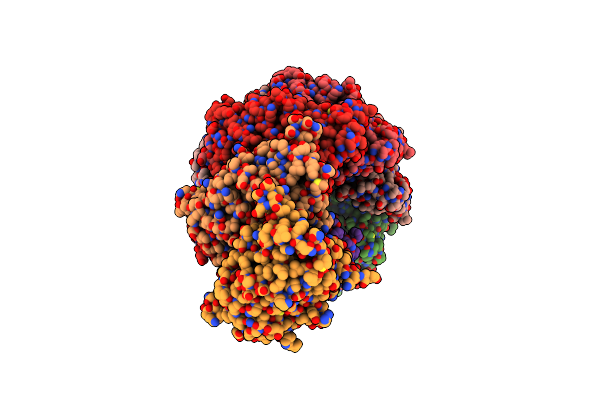 |
Type I-B Cascade Bound To A Pam-Containing Dsdna Target At 3.8 Angstrom Resolution.
Organism: Synechocystis sp. pcc 6714, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: RNA BINDING PROTEIN |
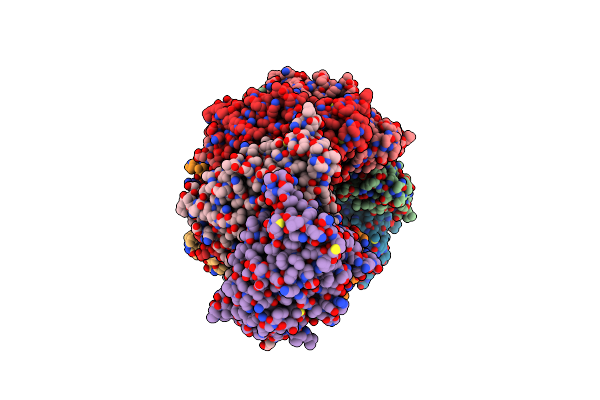 |
Cryo-Em Structure Of Type I-B Cascade Bound To A Pam-Containing Dsdna Target At 3.6 Angstrom Resolution
Organism: Synechocystis sp. pcc 6714, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-03-20 Classification: RNA BINDING PROTEIN/RNA/DNA |
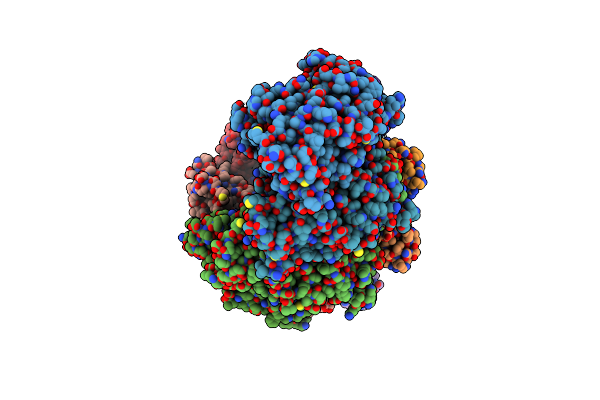 |
Cryo-Em Structure Of Synechocystis Sp. Pcc6714 Cascade At 3.8 Angstrom Resolution
Organism: Synechocystis sp. pcc 6714, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: RNA BINDING PROTEIN |
 |
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: CA, MN, EDO, PEG, PGE, MPD, IMD |
 |
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-05-24 Classification: HYDROLASE Ligands: EDO, ACT, CA, MN, 1PE, PGE, NA, PEG, CL |