Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
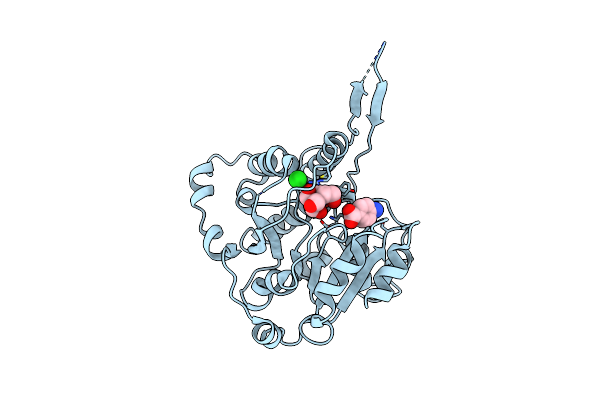
Crystal Structure Of M. Hassiacum Gpgs Co-Crystallized With Udp-Glucose (Ph 7.2)
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Release Date: 2025-09-24 Classification: TRANSFERASE Ligands: TLA, UDP |
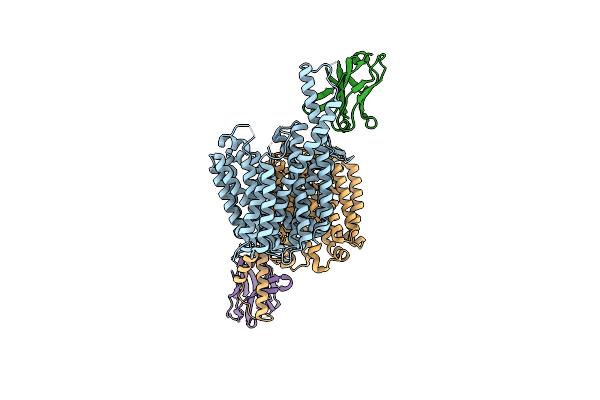 |
Outward-Open Conformation Of A Major Facilitator Superfamily (Mfs) Transporter Mhas2168, A Homologue Of Rv1410 From M. Tuberculosis, In Complex With An Alpaca Nanobody
Organism: Mycolicibacterium hassiacum dsm 44199, Vicugna pacos
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-10-18 Classification: TRANSPORT PROTEIN |
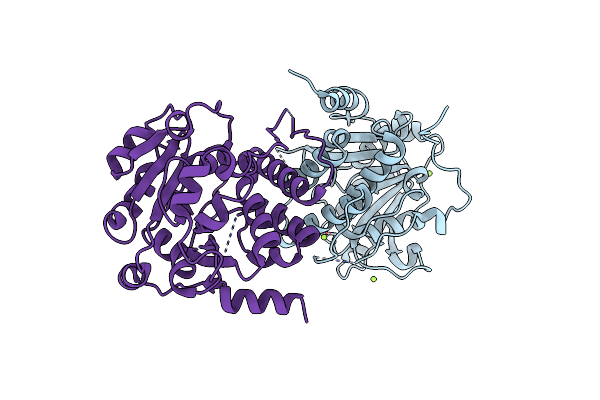 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2023-04-19 Classification: TRANSFERASE Ligands: CL, MG |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.1 In Complex With 4-Hydroxybenzaldehyde
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.27 Å Release Date: 2023-03-01 Classification: TRANSFERASE Ligands: HBA, BCT, GOL, MLT |
 |
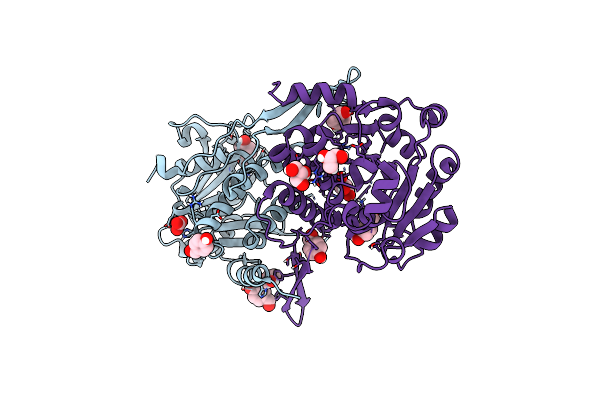
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With Udp-Glucose
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-02-22 Classification: TRANSFERASE Ligands: UPG, MG, CL, BCT |
 |
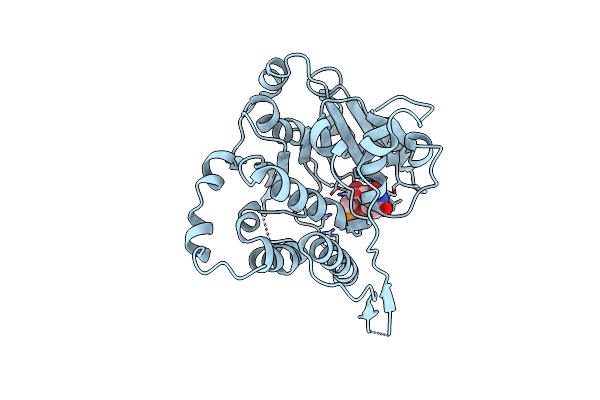
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With 4-Aminobenzoic Acid
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: PAB, BTB, CL |
 |
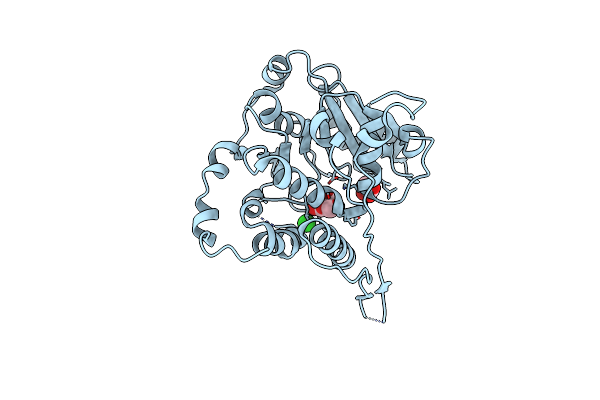
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 In Complex With Udp
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2023-02-15 Classification: TRANSFERASE Ligands: CL, UDP |
 |
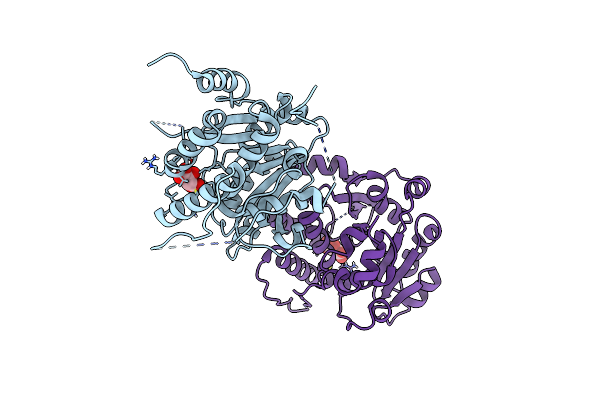
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 5.5 - Apo Form
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2023-01-25 Classification: TRANSFERASE Ligands: CL, BGC, MLI |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.1 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.22 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: LMR, MLT |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 In Complex With Ump And Magnesium
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: MG, U5P, CL |
 |
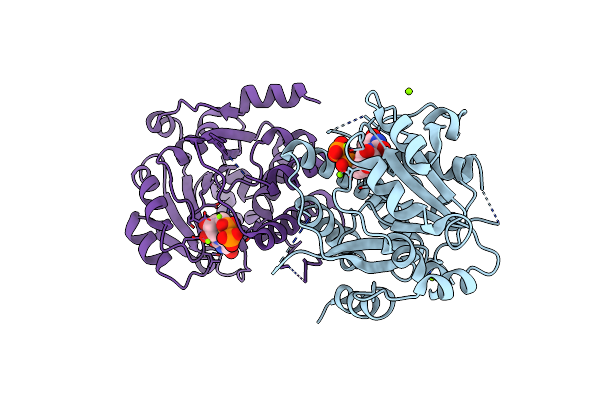
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.2 In Complex With Udp
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: UDP |
 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 8.5 In Complex With Udp
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-12-28 Classification: TRANSFERASE Ligands: MG, UDP, CL |
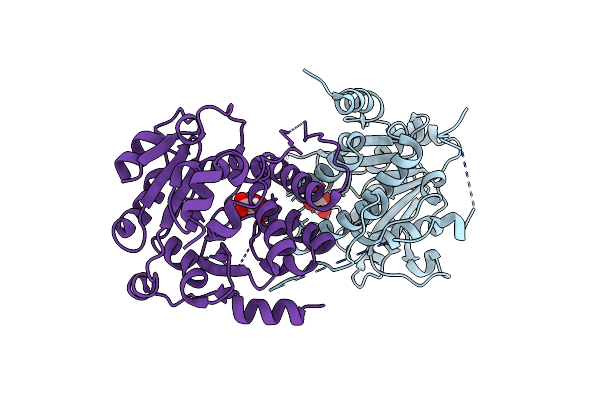 |
Crystal Structure Of Mycobacterium Hassiacum Glucosyl-3-Phosphoglycerate Synthase At Ph 7.2 - Apo Form
Organism: Mycolicibacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.11 Å Release Date: 2022-12-14 Classification: TRANSFERASE Ligands: TLA |
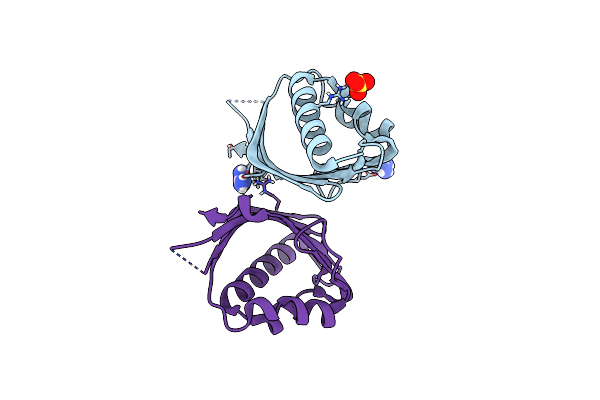 |
Crystal Structure Of Ketosteroid Isomerase From Mycobacterium Hassiacum (Mhksi)
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-05-27 Classification: ISOMERASE Ligands: SO4, GAI |
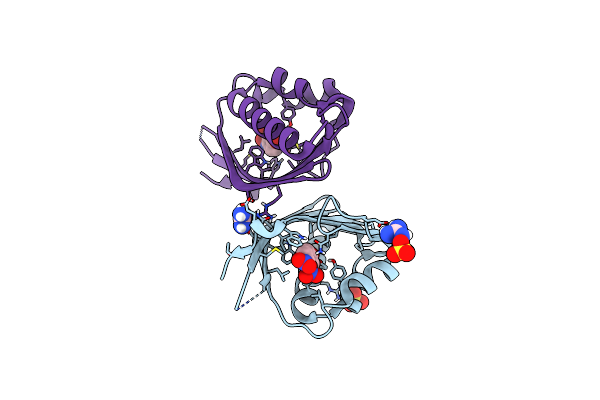 |
Crystal Structure Of Ketosteroid Isomerase D38N Mutant From Mycobacterium Hassiacum (Mhksi) Bound To 3,4-Dinitrophenol
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-05-27 Classification: ISOMERASE Ligands: DNX, SO4, GAI |
 |
Organism: Mycolicibacterium hassiacum dsm 44199
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-12-04 Classification: VIRUS LIKE PARTICLE Ligands: SO4, GOL |
 |
Crystal Structure Of Mycolicibacterium Hassiacum Glucosylglycerate Hydrolase (Mhggh) - Apo Form
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2019-05-15 Classification: HYDROLASE |
 |
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: GOL, SER |
 |
Structure Of Met1 From Mycobacterium Hassiacum In Complex With Sam And Glycerol.
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH, GOL, MG, CL |
 |
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Release Date: 2019-01-16 Classification: TRANSFERASE Ligands: SAH, GOL |