Search Count: 1,697
 |
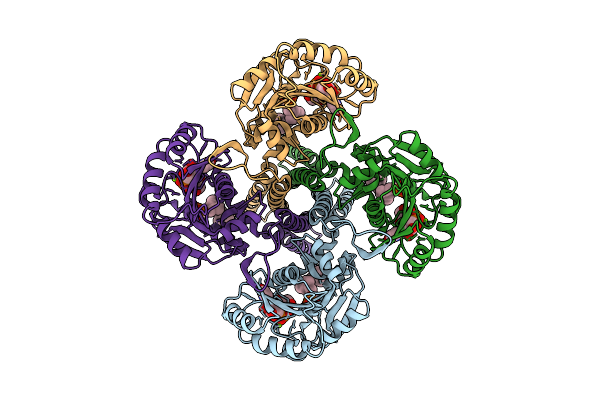
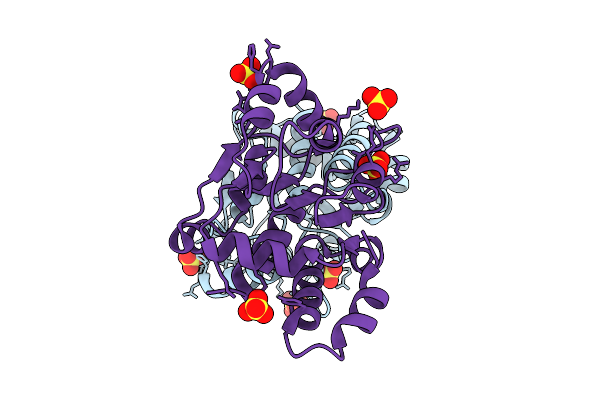
Crystal Structure Of A Putative Phosphate Binding Protein From Synechocystis Sp. Pcc 6803 Reveals An Evolutionary Hotspot
Organism: Synechocystis sp. (strain atcc 27184 / pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: PROTEIN TRANSPORT Ligands: GOL, PO4 |
 |
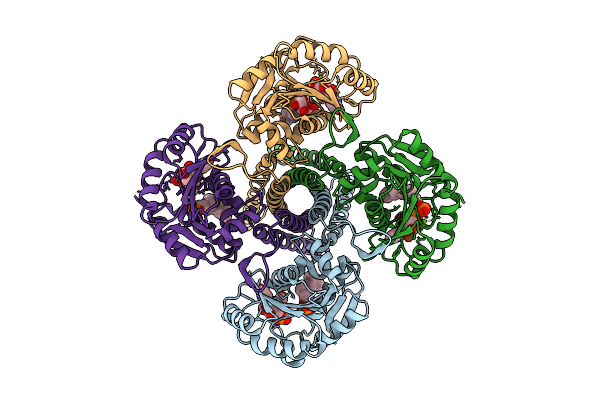
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
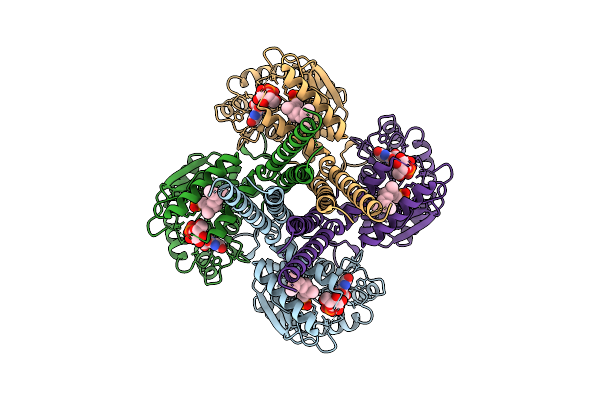
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
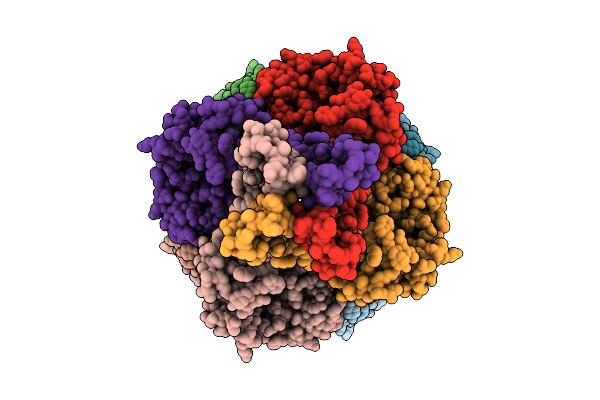 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of Bicarbonate Transporter Sbta In Complex With Pii-Like Signaling Protein Sbtb (T-Loop Truncation) From Synechocystis Sp. Pcc 6803
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSPORT PROTEIN Ligands: BCT |
 |
The C103F Mutant Of Apo-C-Terminal Domain Homolog Of The Orange Carotenoid Protein (Ctdh) From Anabaena
Organism: Cyanobacteriota
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: PHOTOSYNTHESIS Ligands: MLT, FLC |
 |
Organism: Aspergillus fischeri nrrl 181
Method: SOLUTION NMR Release Date: 2025-07-23 Classification: ANTIMICROBIAL PROTEIN |
 |
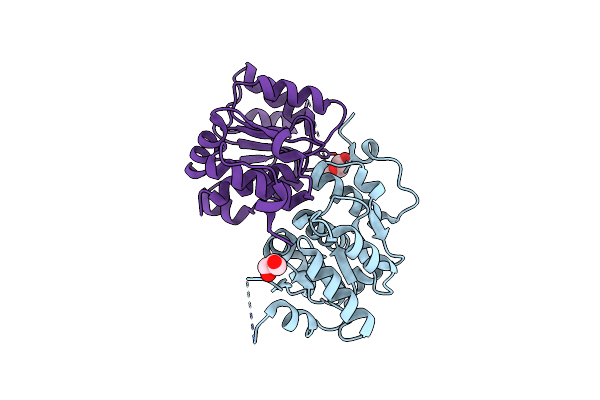
A Tetrapyrrole Binding Domain Variant Of Coar In Closed Conformation At 2.28 Angstrom Resolution
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: METAL BINDING PROTEIN Ligands: SO4 |
 |
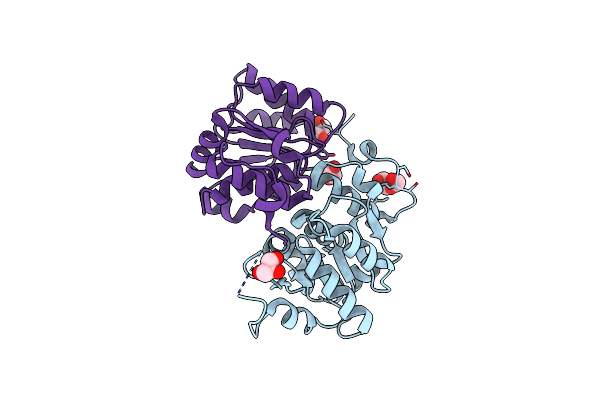
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: METAL BINDING PROTEIN Ligands: GOL, PEG |
 |
Crystal Structure Of The Tetrapyrrole Binding Domain In Coar At 2 Angstrom Resolution
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: METAL BINDING PROTEIN Ligands: PEG, GOL |
 |
Organism: Elizabethkingia anophelis ag1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TOXIN Ligands: CA |
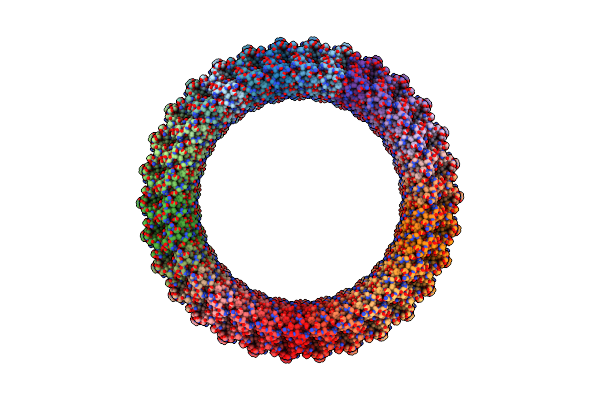 |
Organism: Elizabethkingia anophelis ag1
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: TOXIN Ligands: CA |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: CHAPERONE Ligands: IOD |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: CHAPERONE |
 |
Crystal Structure Of Ccms-Ccmk1-Ccmk2 Complex From Synechocystis Sp. Pcc 6803
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: X-RAY DIFFRACTION Release Date: 2025-03-12 Classification: CHAPERONE Ligands: TAR |
 |
Organism: Synechocystis sp. (strain atcc 27184 / pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2025-01-29 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, IOD |
 |
Organism: Synechocystis sp. (strain pcc 6803 / kazusa)
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: HEM |
 |
Organism: Synechocystis sp. (strain pcc 6803 / kazusa), Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: NI, CL, HEC, GOL |

