Search Count: 141
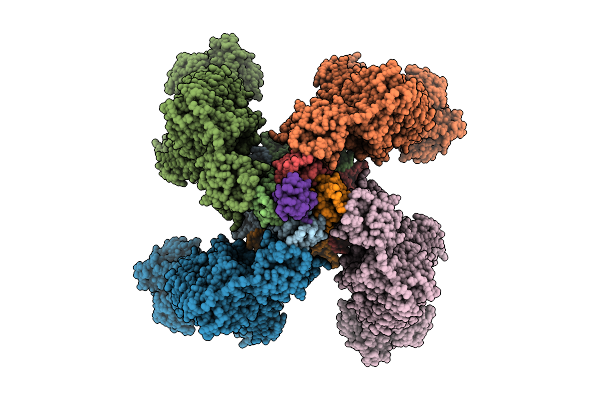 |
Organism: Homo sapiens, Mesocricetus auratus, Scolopendra polymorpha
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: TRANSPORT PROTEIN Ligands: ATP, GBM |
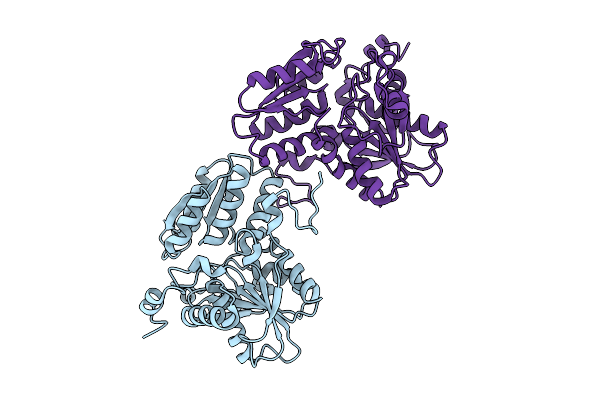 |
Organism: Capnocytophaga ochracea dsm 7271
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN |
 |
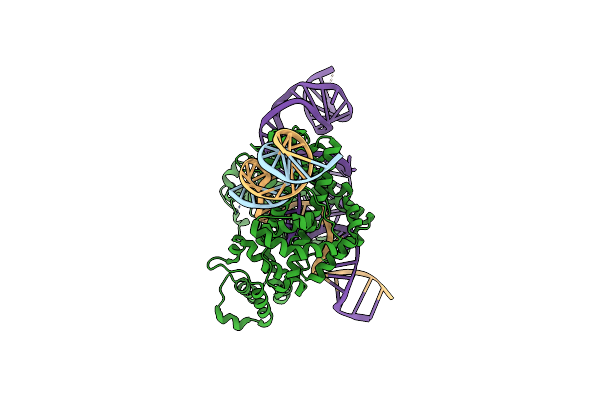
Organism: Escherichia coli k-12, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: ZN |
 |
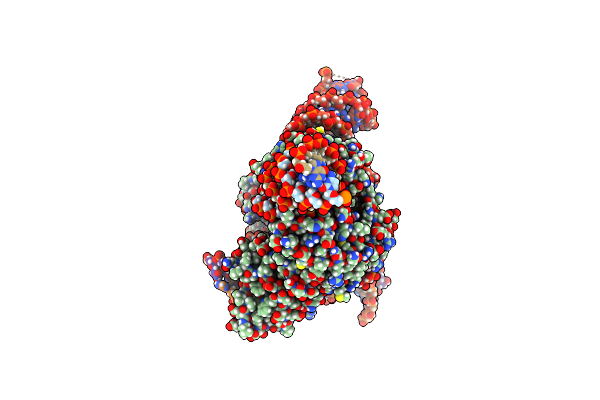
Organism: Escherichia coli k-12, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
 |
Organism: Escherichia coli k-12, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
 |
Organism: Escherichia coli k-12, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: ZN, MG |
 |
Organism: Escherichia coli k-12, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
 |
Organism: Escherichia coli k-12, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-09-11 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
 |
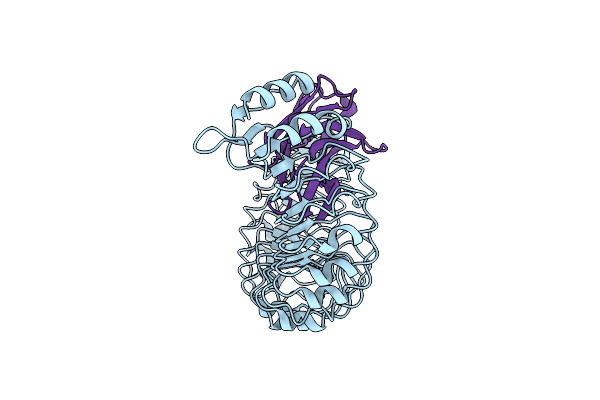
Roco Protein From C. Tepidum In The Gtp State Bound To The Activating Nanobodies Nbroco1 And Nbroco2
Organism: Chlorobaculum tepidum, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: HYDROLASE Ligands: GSP |
 |
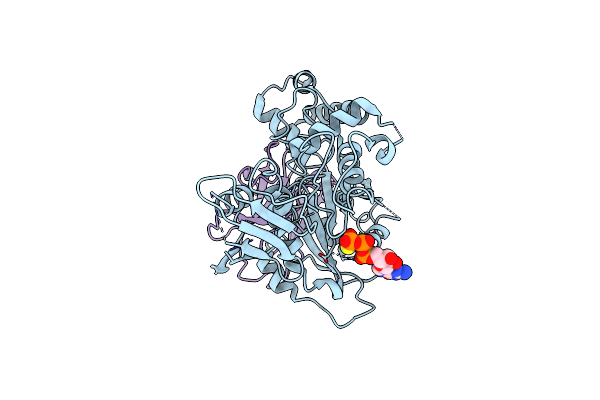
Lrr Domain Of Roco Protein From C. Tepidum Bound To The Activating Nanobody Nbroco2
Organism: Lama glama, Chlorobaculum tepidum
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: HYDROLASE |
 |
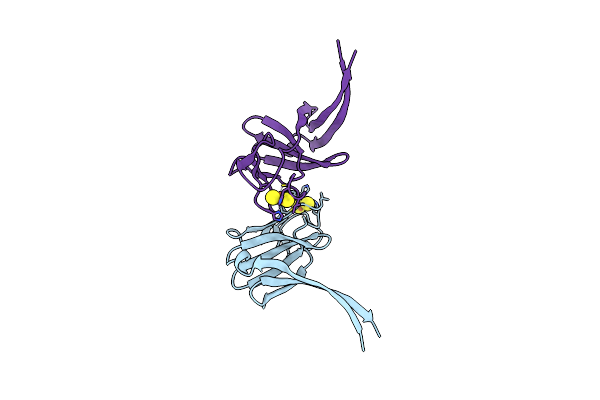
Focused Map On The Roc-Cor Domains Of The Roco Protein From C. Tepidum In The Gtp State Bound To The Activating Nanobody Nbroco1
Organism: Chlorobaculum tepidum, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: HYDROLASE Ligands: GSP |
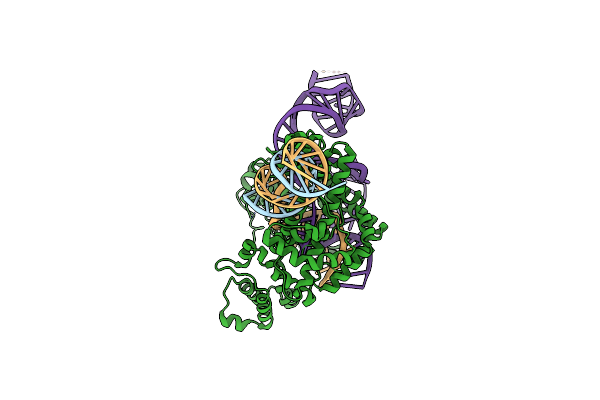 |
Structure Of The Spizellomyces Punctatus Fanzor (Spufz) In Complex With Omega Rna And Target Dna
Organism: Methanosarcina mazei, Spizellomyces punctatus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-07-05 Classification: RNA BINDING PROTEIN/RNA/DNA Ligands: MG, ZN |
 |
Selenomethionine-Labelled Soluble Domain Of Rieske Iron-Sulfur Protein From Chlorobaculum Tepidum
Organism: Chlorobaculum tepidum
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-05 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Organism: Chlorobaculum tepidum
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-07-05 Classification: ELECTRON TRANSPORT Ligands: HEC, SO4, GOL, ACT |
 |
Organism: Streptomyces sp. bc16019
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: SO4, NA |
 |
Organism: Streptomyces sp. bc16019
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: MQW |
 |
Organism: Streptomyces sp. bc16019
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: MQH |
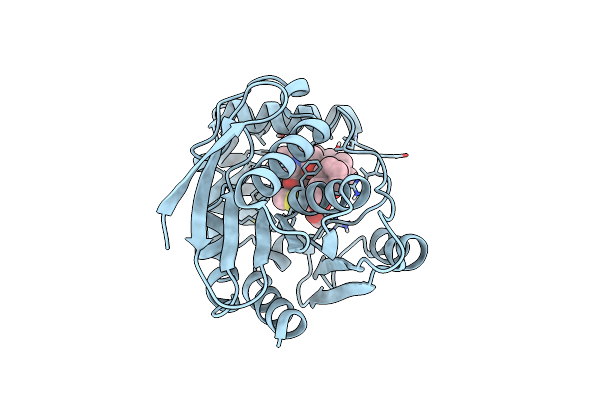 |
Structure Of The Bottromycin Epimerase Both In Complex With A Bottromycin A2 Derivative
Organism: Streptomyces sp. bc16019
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: MRB |
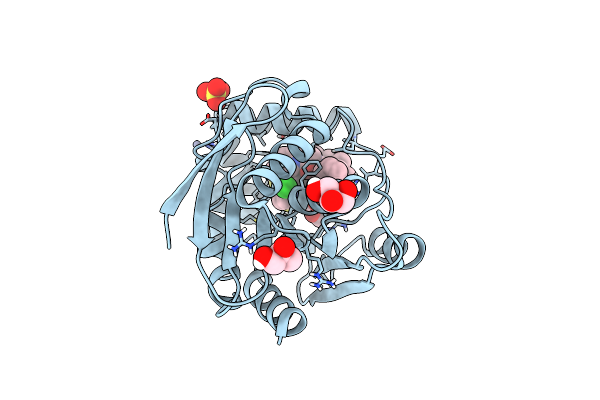 |
Structure Of The Bottromycin Epimerase Both In Complex With Bottromycin A2 Derivative
Organism: Streptomyces sp. bc16019
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2020-07-15 Classification: HYDROLASE Ligands: CL, GOL, SO4, ZN, MQZ |
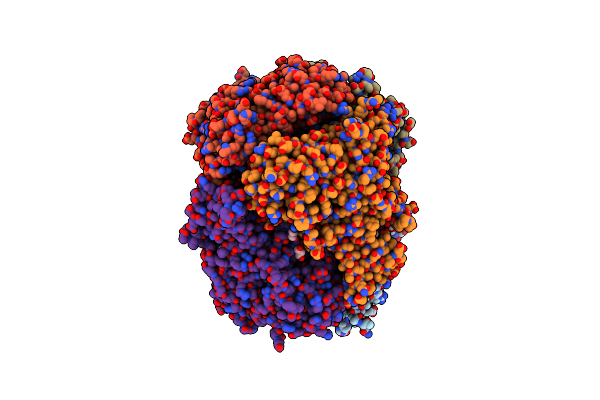 |
Crystal Structure Of Piee, The Flavin-Dependent Monooxygenase Involved In The Biosynthesis Of Piericidin A1
Organism: Streptomyces sp. scsio 03032
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2020-03-11 Classification: FLAVOPROTEIN Ligands: FAD, PGE, 1PE, CL, GOL |

