Search Count: 37
 |
Organism: Mus musculus, Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Resolution:3.93 Å Release Date: 2018-11-21 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Organism: Mus musculus, Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Resolution:3.93 Å Release Date: 2018-11-21 Classification: IMMUNE SYSTEM Ligands: ZN |
 |
Atomic Structure Fitted Into A Localized Reconstruction Of Bacteriophage Phi6 Packaging Hexamer P4
Organism: Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Release Date: 2017-03-22 Classification: HYDROLASE Ligands: CA, ADP |
 |
Atomic Structure Of P4 Packaging Enzyme Fitted Into A Localized Reconstruction Of Bacteriophage Phi6 Vertex
Organism: Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Resolution:9.10 Å Release Date: 2017-03-22 Classification: HYDROLASE Ligands: CA, ADP |
 |
Organism: Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Resolution:4.80 Å Release Date: 2015-11-04 Classification: VIRAL PROTEIN |
 |
Structure Of The P2 Polymerase Inside In Vitro Assembled Bacteriophage Phi6 Polymerase Complex
Organism: Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Resolution:7.90 Å Release Date: 2015-11-04 Classification: TRANSCRIPTION Ligands: MN |
 |
Structure Of The P2 Polymerase Inside In Vitro Assembled Bacteriophage Phi6 Polymerase Complex, With P1 Included
Organism: Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Resolution:7.90 Å Release Date: 2015-11-04 Classification: VIRAL PROTEIN Ligands: MN |
 |
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-08-21 Classification: HYDROLASE Ligands: CA, ADP |
 |
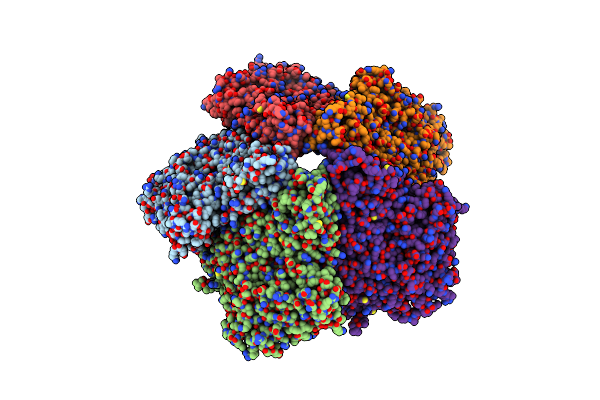
Coordinates Of The Bacteriophage Phi6 Capsid Subunits (P1A And P1B) Fitted Into The Cryoem Reconstruction Of The Procapsid At 4.4 A Resolution
Organism: Pseudomonas phage phi6
Method: ELECTRON MICROSCOPY Resolution:4.40 Å Release Date: 2013-08-14 Classification: VIRUS |
 |
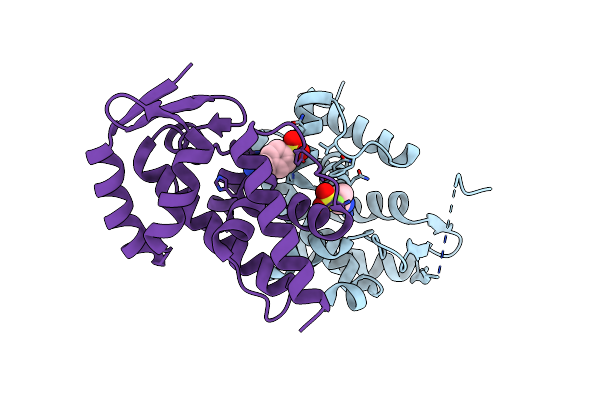
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2013-08-14 Classification: VIRAL PROTEIN |
 |
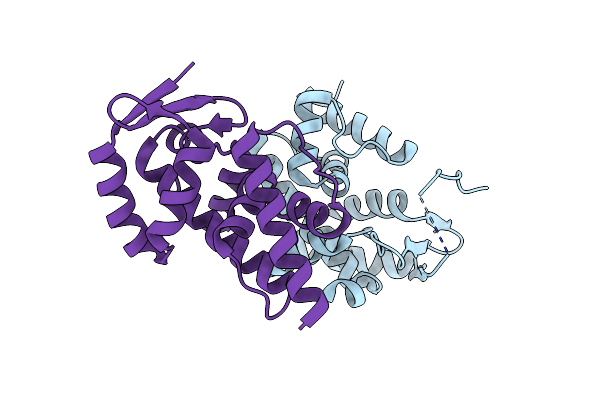
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-01-02 Classification: MEMBRANE PROTEIN Ligands: AES |
 |
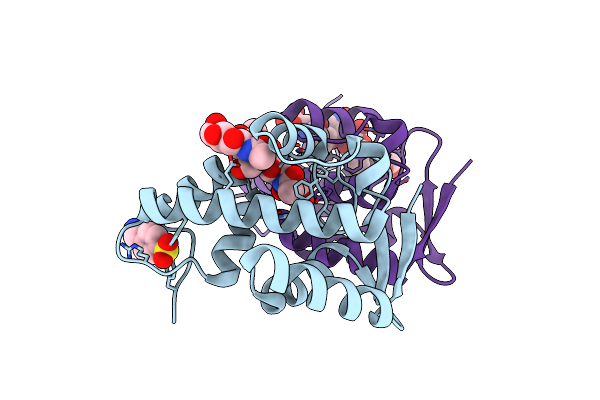
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2013-01-02 Classification: HYDROLASE |
 |
Structural Investigation Of Bacteriophage Phi6 Lysin (In Complex With Chitotetraose)
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2013-01-02 Classification: HYDROLASE Ligands: AES |
 |
The C-Terminal Priming Domain Is Strongly Associated With The Main Body Of Bacteriophage Phi6 Rna-Dependent Rna Polymerase
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-08-01 Classification: TRANSFERASE Ligands: MN |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: MG, ATP |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2012-07-04 Classification: TRANSFERASE/DNA Ligands: MG, GTP |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2012-07-04 Classification: TRANSFERASE/DNA Ligands: MG, GTP, ATP |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2012-07-04 Classification: TRANSFERASE Ligands: MN |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6
Method: X-RAY DIFFRACTION Resolution:3.06 Å Release Date: 2012-07-04 Classification: TRANSFERASE/DNA Ligands: MG, ATP |
 |
Non-Catalytic Ions Direct The Rna-Dependent Rna Polymerase Of Bacterial Dsrna Virus Phi6 From De Novo Initiation To Elongation
Organism: Pseudomonas phage phi6, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-07-04 Classification: TRANSFERASE/DNA Ligands: MG, ATP |

