Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Rhodospirillum rubrum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: OXIDOREDUCTASE Ligands: S5Q, CLF, ADP, MG, AF3, SF4 |
 |
Crystal Structure Of The Recombinant Codh From Rhodopspirillum Rubrum Produced In Escherichia Coli
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2025-02-19 Classification: OXIDOREDUCTASE Ligands: SF4, RQM, NA |
 |
Organism: Rhodospirillum rubrum
Method: ELECTRON MICROSCOPY Release Date: 2024-12-04 Classification: PHOTOSYNTHESIS Ligands: 07D, CRT, FE, BPH, U10 |
 |
Organism: Spiromicrovirus spv4
Method: ELECTRON MICROSCOPY Release Date: 2024-07-17 Classification: VIRUS |
 |
Organism: Chromobacterium vaccinii
Method: X-RAY DIFFRACTION Release Date: 2023-07-12 Classification: OXIDOREDUCTASE Ligands: ACT, GOL, O, FE |
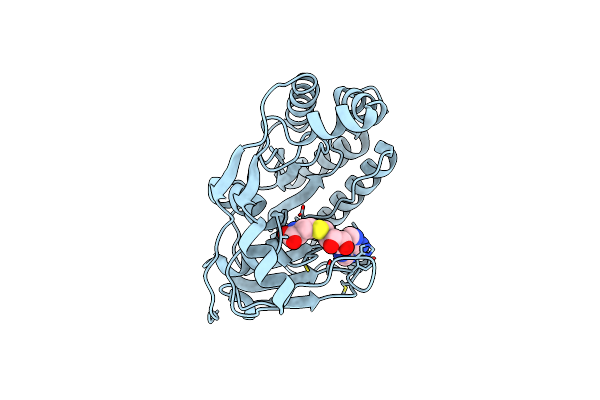 |
Crystal Structure And Cap Binding Analysis Of The Methyltransferase Of Langat Virus
Organism: Langat virus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-01-25 Classification: STRUCTURAL PROTEIN Ligands: SAH |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE Ligands: FE, CA |
 |
Organism: Rhodospirillum rubrum
Method: SOLUTION NMR Release Date: 2020-03-18 Classification: METAL BINDING PROTEIN |
 |
Organism: Rhodospirillum rubrum
Method: SOLUTION NMR Release Date: 2020-03-18 Classification: METAL BINDING PROTEIN |
 |
X-Ray Structure Of A Truncated Mutant Of The Metallochaperone Cooj With A High-Affinity Nickel-Binding Site
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2019-03-27 Classification: METAL BINDING PROTEIN Ligands: NI, PEG, CL, CA, PGE, Z3P |
 |
Crystal Structure Of The Apo-Form Of The Co Dehydrogenase Accessory Protein Coot From Rhodospirillum Rubrum
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-05-10 Classification: nickel-binding protein |
 |
Structure Function Studies Of R. Palustris Rubisco (R. Palustris/R. Rubrum Chimera)
Organism: Rhodopseudomonas palustris, Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-01-25 Classification: LYASE Ligands: CAP, MG, K |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2016-08-31 Classification: OXIDOREDUCTASE Ligands: CA |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2016-08-10 Classification: OXIDOREDUCTASE Ligands: FE, GOA, CA |
 |
Organism: Burkholderia pseudomallei 1026a
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2012-12-12 Classification: TOXIN Ligands: BR |
 |
Crystal Structure Of The D97N Variant Of Dinitrogenase Reductase- Activating Glycohydrolase (Drag) From Rhodospirillum Rubrum In Complex With Adp-Ribose
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-08-18 Classification: HYDROLASE Ligands: MN, AR6, TLA, GOL |
 |
Crystal Structure Of The Dinitrogenase Reductase-Activating Glycohydrolase (Drag) From Rhodospirillum Rubrum
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-08-11 Classification: HYDROLASE Ligands: MN, FMT, GOL, CL |
 |
Crystal Structure Of The Dinitrogenase Reductase-Activating Glycohydrolase (Drag) From Rhodospirillum Rubrum In Complex With Adp- Ribsoyllysine
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2009-08-11 Classification: HYDROLASE Ligands: MN, ZZC, CL, GOL |