Search Count: 41
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RECOMBINATION/DNA |
 |
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RECOMBINATION/DNA |
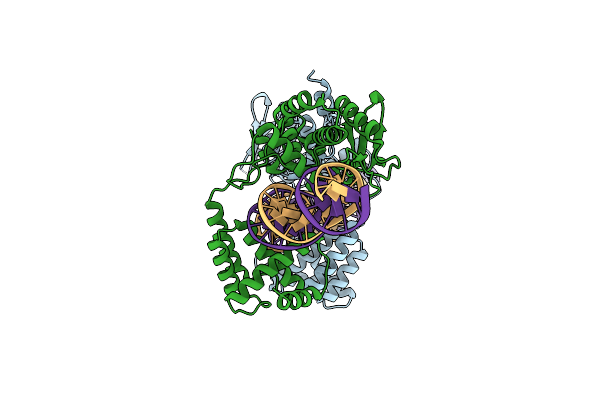 |
Heterodimer Of Cre Recombinase Mutants D33A/A36V/R192A And R72E/L115D/R119D In Complex With Loxp Dna.
Organism: Escherichia phage p1
Method: ELECTRON MICROSCOPY Release Date: 2022-01-19 Classification: RECOMBINATION/DNA |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION Ligands: ACT |
 |
Antitoxin Phd From Phage P1 In Complex With Its Operator Dna Inverted Repeat
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION |
 |
Antitoxin Phd From Phage P1 In Complex With Its Operator Dna Inverted Repeat In A Monoclinic Space Group
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.88 Å Release Date: 2016-04-20 Classification: TRANSCRIPTION |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-12-29 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
Organism: Enterobacteria phage p1, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-08-18 Classification: TOXIN/ANTITOXIN Ligands: ACT |
 |
Crystal Structure Of The P1 Bacteriophage Doc Toxin (F68S) In Complex With The Phd Antitoxin (L17M/V39A). Northeast Structural Genomics Targets Er385-Er386
Organism: Bacteriophage p1
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2010-08-18 Classification: TOXIN Ligands: CL, HED, GOL, PO4 |
 |
Crystal Structure Of Phd Truncated To Residue 57 In An Orthorhombic Space Group
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2010-06-23 Classification: ANTITOXIN Ligands: SO4 |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2010-05-26 Classification: ISOMERASE/DNA Ligands: VO4 |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2009-06-09 Classification: RIBOSOME INHIBITOR |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2008-09-16 Classification: RIBOSOME INHIBITOR Ligands: BR |
 |
Crystal Structure Of The Pre-Cleavage Synaptic Complex In The Cre-Loxp Site-Specific Recombination
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-06-26 Classification: recombination/DNA |
 |
Crystal Structure Of The Tetrameric Pre-Cleavage Synaptic Complex In The Cre-Loxp Site-Specific Recombination
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2007-06-26 Classification: RECOMBINATION/DNA |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2007-02-20 Classification: CELL CYCLE/DNA |
 |
Organism: Escherichia coli, Enterobacteria phage
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2006-11-14 Classification: TRANSFERASE Ligands: MN, TMP, EDO |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2005-11-29 Classification: CELL CYCLE Ligands: CIT |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2005-01-11 Classification: HYDROLASE Ligands: CIT |
 |
Organism: Enterobacteria phage p1
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2005-01-11 Classification: HYDROLASE Ligands: SO4 |

