Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2024-02-28 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-04-21 Classification: LIGASE,TRANSFERASE |
 |
2-Amino-3-Ketobutyrate Coa Ligase From Cupriavidus Necator L-Threonine Binding Form
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-04-21 Classification: LIGASE,TRANSFERASE Ligands: 2BK |
 |
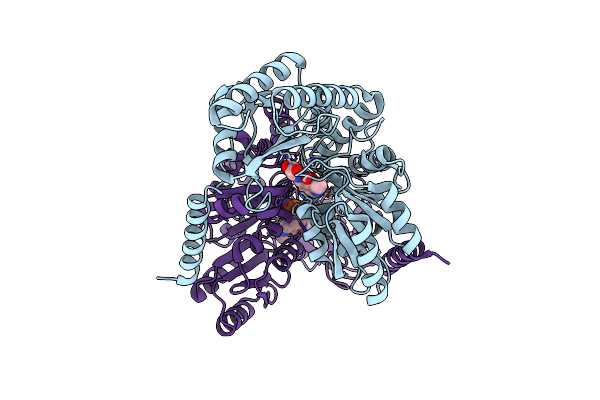
2-Amino-3-Ketobutyrate Coa Ligase From Cupriavidus Necator 3-Hydroxynorvaline Binding Form
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-04-21 Classification: LIGASE,TRANSFERASE Ligands: F9X |
 |
2-Amino-3-Ketobutyrate Coa Ligase From Cupriavidus Necator Glycine Binding Form
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-04-21 Classification: LIGASE,TRANSFERASE Ligands: PLG |
 |
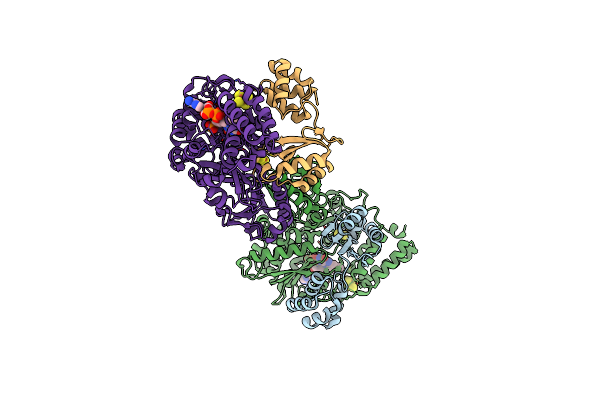
Crystal Structure Of Full-Length Cbnr Complexed With The Target Dna Complex
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2021-03-10 Classification: DNA BINDING PROTEIN/DNA |
 |
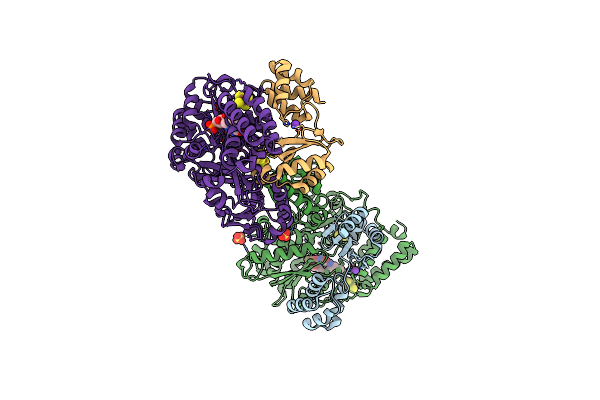
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE Ligands: FES, FMN, SF4, NAI |
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE Ligands: FES, K, FMN, SF4, SO4 |
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2018-01-17 Classification: DNA BINDING PROTEIN |
 |
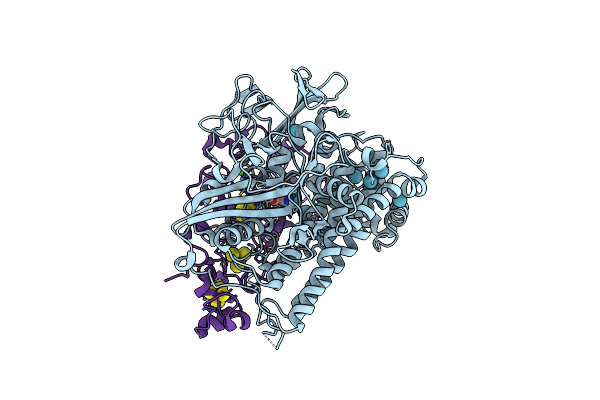
Structure Of The Catalytic Domain Of The Class I Polyhydroxybutyrate Synthase From Cupriavidus Necator
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-26 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
Krypton Derivatization Of An O2-Tolerant Membrane-Bound [Nife] Hydrogenase Reveals A Hydrophobic Gas Tunnel Network
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2016-03-23 Classification: OXIDOREDUCTASE Ligands: NFV, MG, KR, CL, SF4, F3S, F4S |
 |
Crystal Structure Of A Putative 4-Methylmuconolactone Methylisomerase (Yp_295714.1) From Ralstonia Eutropha Jmp134 At 2.00 A Resolution
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-10-10 Classification: Structural Genomics/Unknown function Ligands: CL, GOL |
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2003-06-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2003-05-06 Classification: DNA BINDING PROTEIN |
 |
The Crystal Structure Of Rubisco From Alcaligenes Eutrophus To 2.7 Angstroms.
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 1999-10-06 Classification: LYASE Ligands: PO4 |
 |
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 1995-07-31 Classification: ISOMERASE Ligands: MN, CL |
 |
 |
 |
 |
NA
|