Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
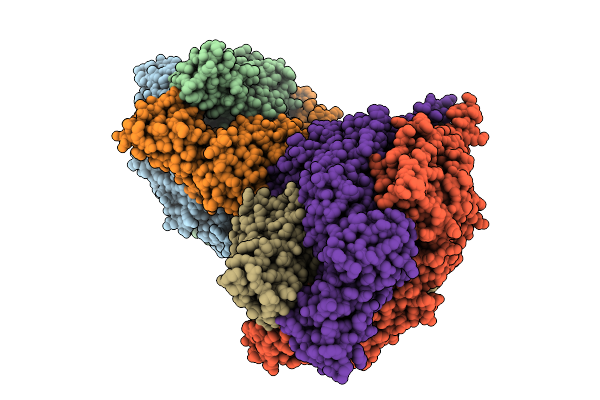
Cyro-Em Structure Of Prefusion Rsv Fusion Glycoprotein In Complex With Ziresovir And Motavizumab Fab
Organism: Human respiratory syncytial virus a2, Tequatrovirus t4, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: A1EV1 |
 |
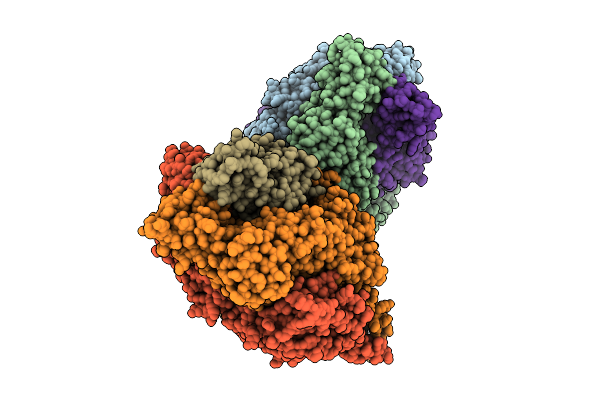
Organism: Human respiratory syncytial virus, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
 |
Organism: Respiratory syncytial virus, Enterobacteria phage t6
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
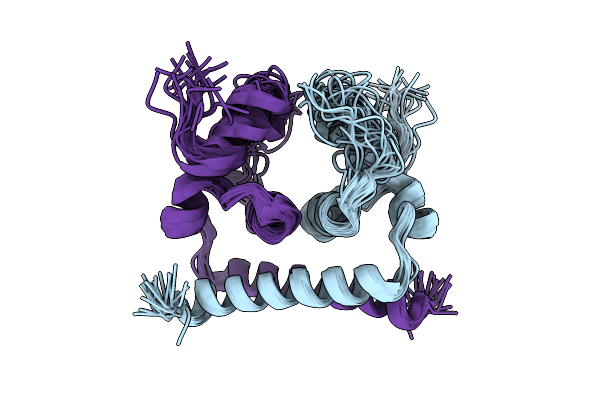 |
Nmr Strucuture Of Dengue Virus 2 Capsid Protein With The Deletion Of The Intrinsically Disordered N-Terminal Region
Organism: Dengue virus type 2
Method: SOLUTION NMR Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
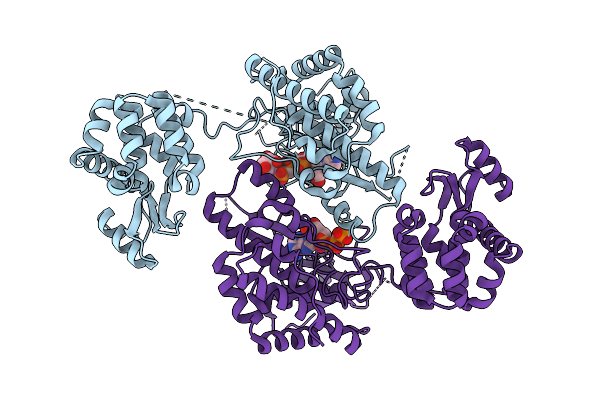 |
Crystal Structure Of Phosphatidyl Inositol 4-Kinase Ii Beta In Complex With Hh5129
Organism: Homo sapiens, Enterobacteria phage t4
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: A1IVA |
 |
Organism: Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: ISOMERASE |
 |
Dengue 2 Virus Ns2B-Ns3 Protease Fusion Protein With Crystal Epitope Mutation K174Q
Organism: Dengue virus type 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: VIRAL PROTEIN |
 |
Organism: Escherichia phage t4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: ISOMERASE Ligands: MG |
 |
Crystal Structure Of The Transmembrane Domain Of Trimeric Autotransporter Adhesin Ataa In Complex With The N-Terminal Domain Of Tpga
Organism: Acinetobacter sp. tol 5
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: CELL ADHESION Ligands: CA, C8E, PGE, MES, CL |
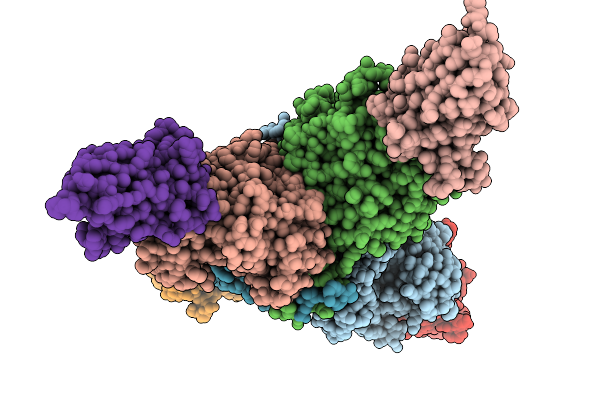 |
Crystal Structure Of The Complex Between Nb474 Mutant R53A,D125A And Trypanosoma Congolense Fructose-1,6-Bisphosphate Aldolase
Organism: Trypanosoma congolense il3000, Vicugna pacos
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: LYASE Ligands: ACT, GOL |
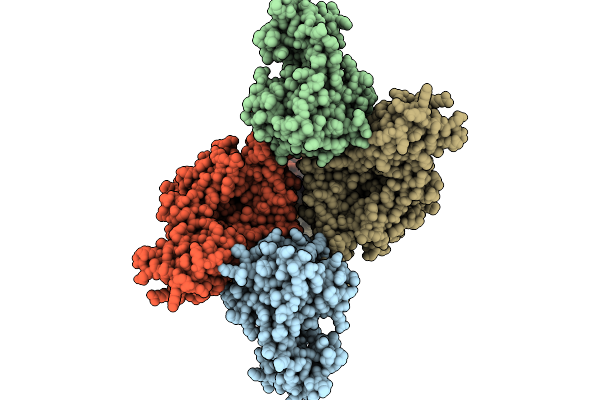 |
Organism: Escherichia phage t4, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: ISOMERASE/DNA |
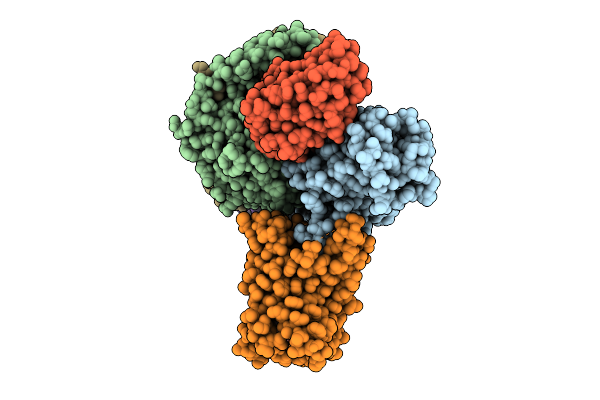 |
Organism: Spodoptera
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: MEMBRANE PROTEIN Ligands: AND |
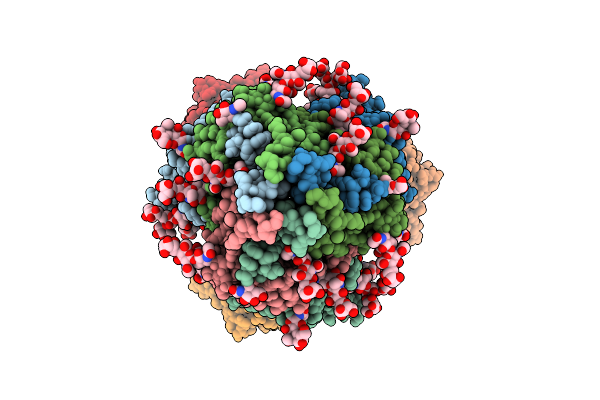 |
Pre-Fusion Herv-K Envelope Protein Trimer Ectodomain In Complex With Kenv-6 Fab
Organism: Homo sapiens, Enterobacteria phage t4, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Enterobacteria phage t4
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Tequatrovirus t4, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-23 Classification: PROTEIN BINDING Ligands: NAG |
 |
Organism: Lachnospiraceae bacterium nd2006, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/RNA |
 |
Organism: Lachnospiraceae bacterium nd2006, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: header DNA BINDING PROTEIN/RNA |
 |
Organism: Escherichia coli uti89, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: CELL ADHESION |
 |
Organism: Escherichia coli uti89, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: CELL ADHESION |
 |
Organism: Escherichia coli uti89, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: CELL ADHESION |