Search Count: 121
 |
Organism: Bat coronavirus hku5, Pipistrellus abramus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Structure Of Hku5 Spike C-Terminal Domain In Complex With Ace2 From Pipistrellus Abramus
Organism: Pipistrellus abramus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Klebsormidium nitens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-02 Classification: ELECTRON TRANSPORT Ligands: PC1, RET |
 |
Organism: Pipistrellus abramus, Pipistrellus bat coronavirus hku5
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-02-19 Classification: VIRAL PROTEIN/HYDROLASE Ligands: NAG, ZN |
 |
The Crystal Structure Of Pet44, A Petase Enzyme From Alkalilimnicola Ehrlichii
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-11-06 Classification: HYDROLASE |
 |
Crystal Structure Of The Lov1 R60K Mutant Of Klebsormidium Nitens Phototropin
Organism: Klebsormidium nitens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Crystal Structure Of Lov1 D33N Mutant Of Phototropin From Klebsormidium Nitens
Organism: Klebsormidium nitens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-04-10 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Crystal Structure Of The Lov1 Q122N Mutant Of Klebsormidium Nitens Phototropin
Organism: Klebsormidium nitens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2024-03-06 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Organism: Klebsormidium nitens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-01-24 Classification: FLAVOPROTEIN Ligands: FMN |
 |
Organism: Human adenovirus f serotype 41
Method: ELECTRON MICROSCOPY Release Date: 2020-11-25 Classification: VIRAL PROTEIN |
 |
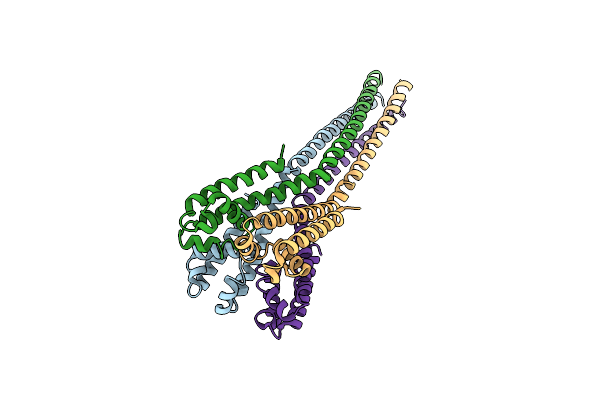
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:3.75 Å Release Date: 2016-03-30 Classification: TRANSPORT PROTEIN Ligands: BR |
 |
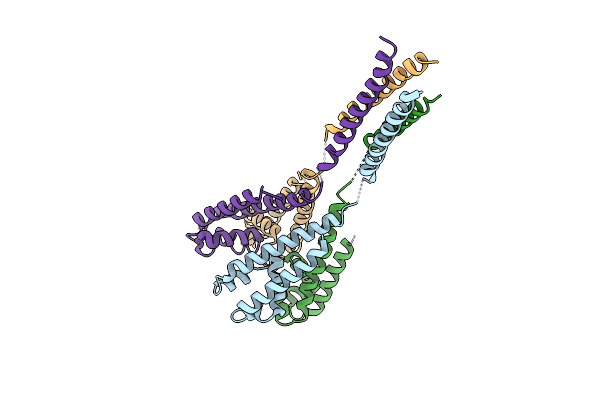
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2016-03-30 Classification: TRANSPORT PROTEIN Ligands: BR |
 |
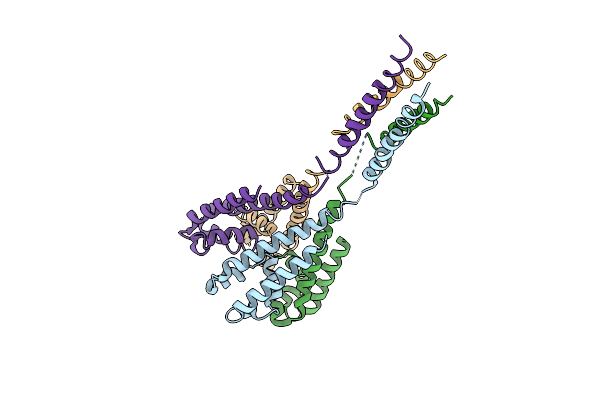
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
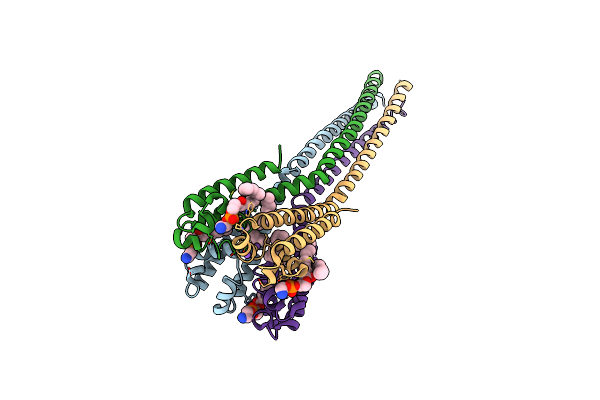
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN |
 |
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN Ligands: CL, VVA, NA |
 |
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2016-03-09 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Crystal Structure Of Purine Nucleoside Phosphorylase From Streptococcus Agalactiae 2603V/R, Nysgrc Target 030935
Organism: Streptococcus agalactiae lmg 15084
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-07-23 Classification: TRANSFERASE Ligands: SO4 |
 |
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2013-10-23 Classification: TRANSPORT PROTEIN Ligands: CA, NI |
 |
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:3.80 Å Release Date: 2013-10-23 Classification: TRANSPORT PROTEIN Ligands: CA |
 |
Organism: Alkalilimnicola ehrlichii
Method: X-RAY DIFFRACTION Resolution:5.50 Å Release Date: 2013-10-23 Classification: TRANSPORT PROTEIN |

