Search Count: 98
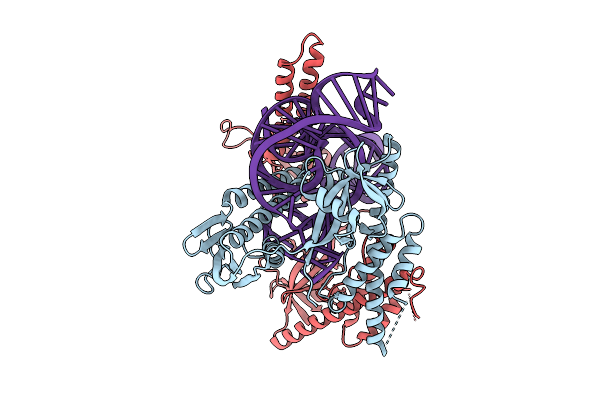 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
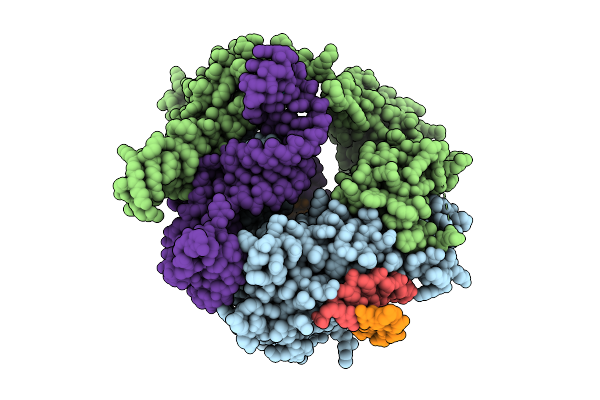 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA |
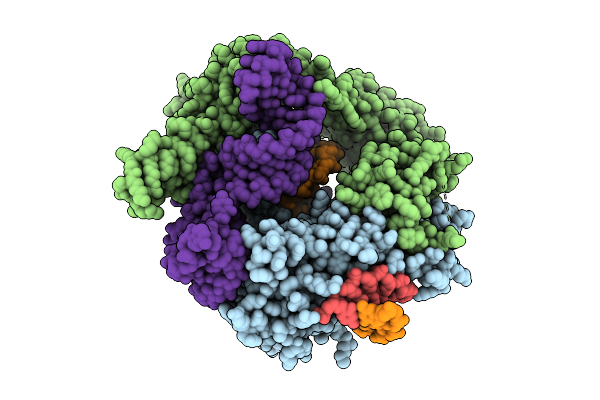 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA |
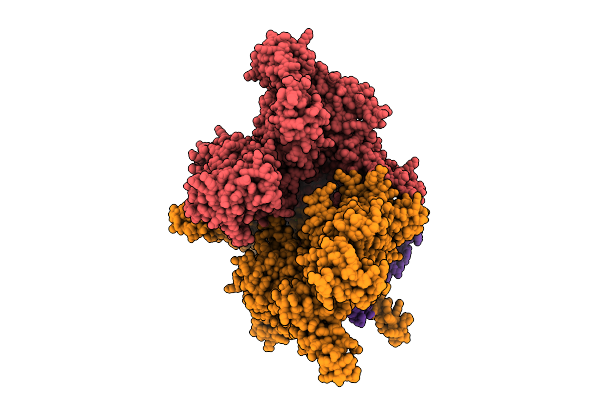 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
 |
Organism: Escherichia coli 0.1197
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2025-02-19 Classification: DNA BINDING PROTEIN |
 |
Organism: Escherichia coli, Escherichia coli 0.1197
Method: ELECTRON MICROSCOPY Release Date: 2025-02-05 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Ogataea parapolymorpha
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-09-04 Classification: HYDROLASE Ligands: GOL, MG |
 |
Organism: Escherichia coli, Escherichia coli 0.1288, Escherichia coli o157
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: MEMBRANE PROTEIN/Hydrolase |
 |
Contact-Dependent Growth Inhibition Toxin-Immunity Protein Complex From From E. Coli 3006, Full-Length
Organism: Escherichia coli 3006
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2021-01-27 Classification: TOXIN |
 |
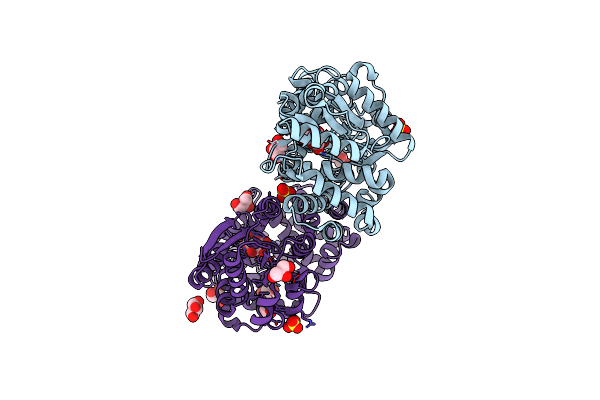
Crystal Structure Of Periplasmic Glucose Binding Protein Ppgbp Deletion Mutant- Del-Ppgbp
Organism: Pseudomonas putida csv86
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-10-10 Classification: TRANSPORT PROTEIN |
 |
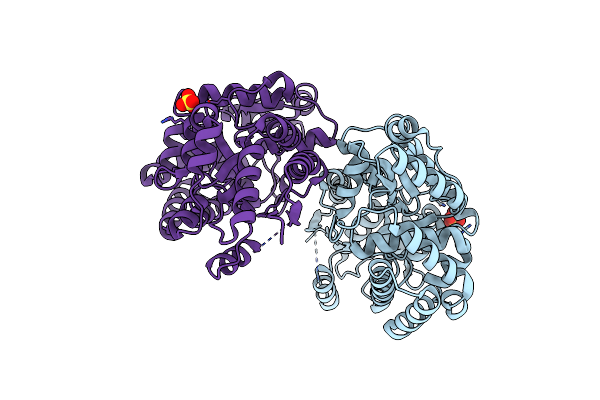
High Resolution Crystal Structure Of Glucose Complexed Periplasmic Glucose Binding Protein (Ppgbp) From P. Putida Csv86
Organism: Pseudomonas putida csv86
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2016-02-24 Classification: TRANSPORT PROTEIN Ligands: BGC, GOL, SO4 |
 |
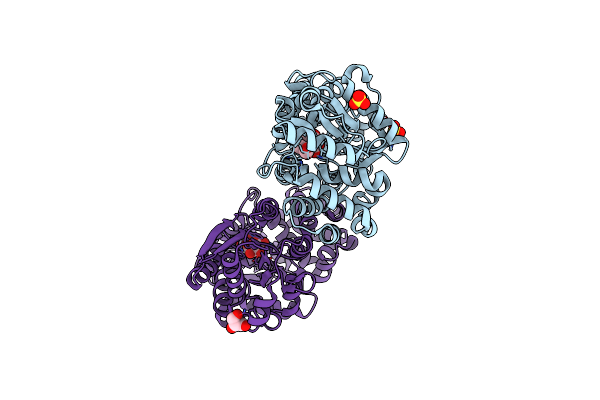
Crystal Structure Of Unliganded Periplasmic Glucose Binding Protein (Ppgbp) From P. Putida Csv86
Organism: Pseudomonas putida csv86
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-02-17 Classification: TRANSPORT PROTEIN Ligands: SO4 |
 |
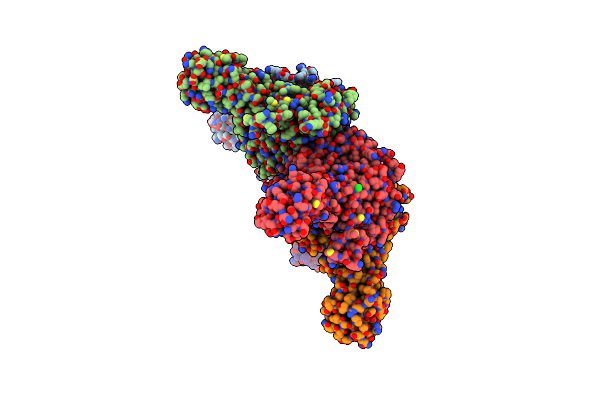
Crystal Structure Of Galactose Complexed Periplasmic Glucose Binding Protein (Ppgbp) From P. Putida Csv86
Organism: Pseudomonas putida csv86
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-02-17 Classification: TRANSPORT PROTEIN Ligands: GAL, GOL, SO4 |
 |
Organism: Infectious pancreatic necrosis virus -
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2009-12-29 Classification: VIRUS LIKE PARTICLE Ligands: CO, CL |
 |
Crystal Structure Of Vp4 Protease From Infectious Pancreatic Necrosis Virus (Ipnv) In Space Group P6122
Organism: Infectious pancreatic necrosis virus - sp
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-06-05 Classification: HYDROLASE |
 |
 |
 |
 |

