Search Count: 19,992
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
 |
Mycobacterium Tuberculosis Relbe1 Toxin-Antitoxin System; Rv1247C (Relb1 Antitoxin), Rv1246C (Rele1 Toxin)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TOXIN Ligands: CL |
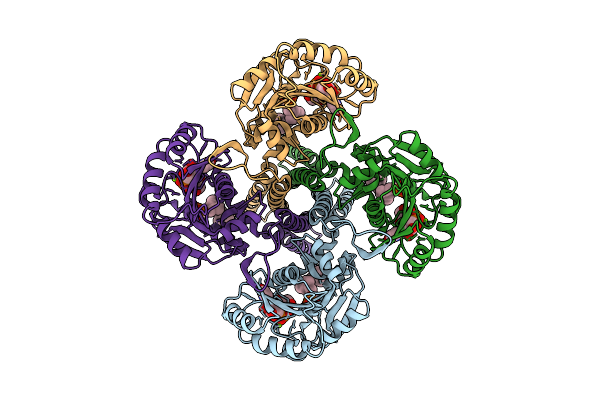 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
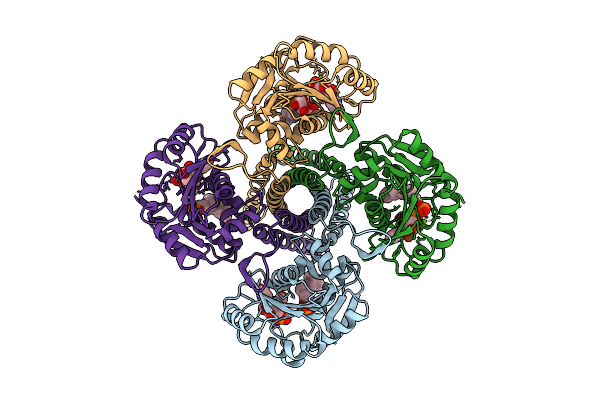 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
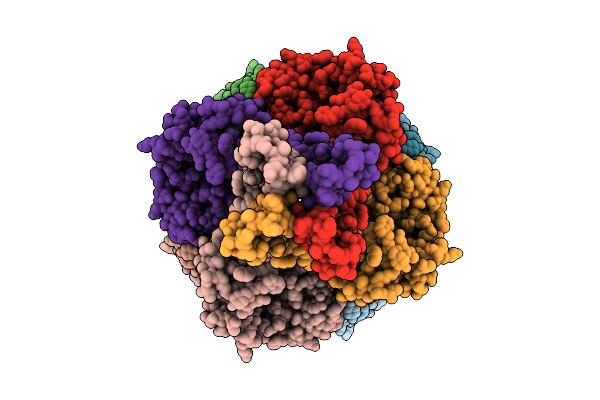 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
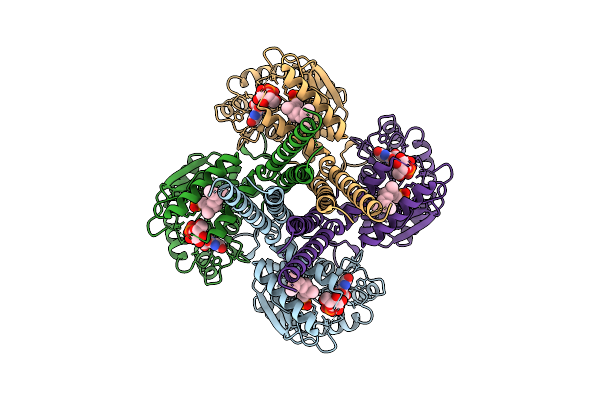
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1CIF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1AZ4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1CIF |
 |
Organism: Human adenovirus d10
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRUS |
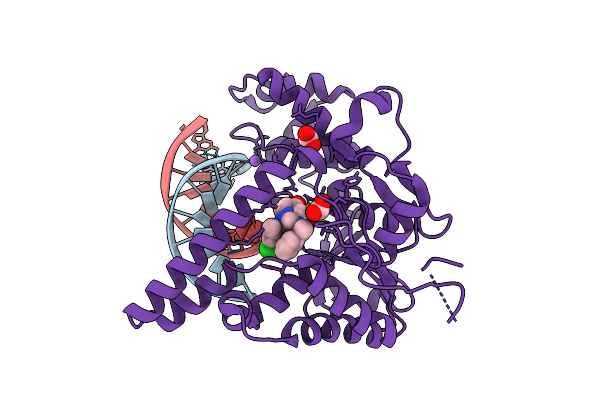 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: MG, NA, A1JNC, FMT |
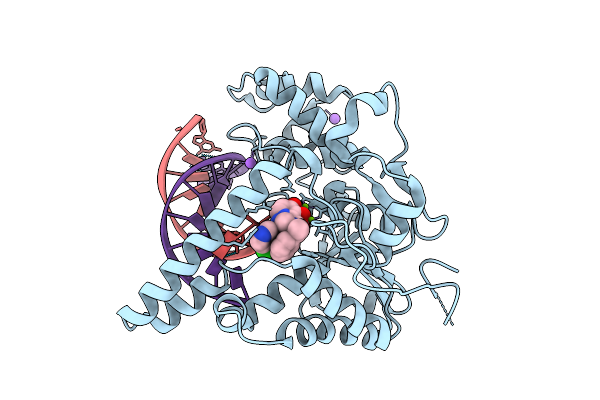 |
Crystal Structure Of Human Exonuclease 1 (Exo1) With Dna And Compound Art5537
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: HYDROLASE Ligands: MG, A1JO0, NA |
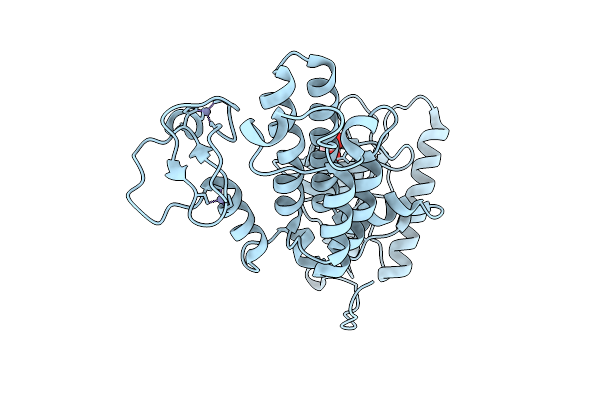 |
Crystal Structure Of Human Prkcbp1 Zinc Finger Mynd-Type Containing 8 With Pentaglutamate Tag
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: EDO, ZN |
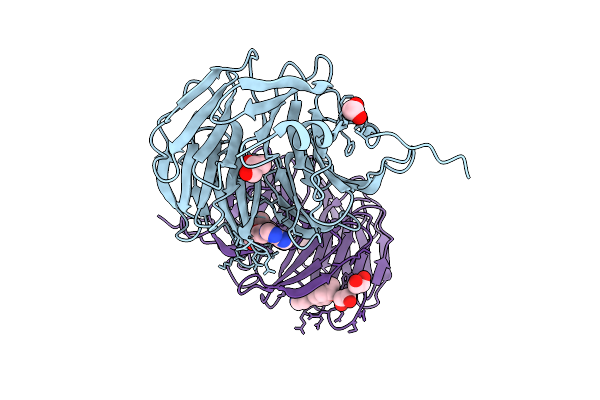 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: NUCLEAR PROTEIN Ligands: GOL, A1BI5, EDO |
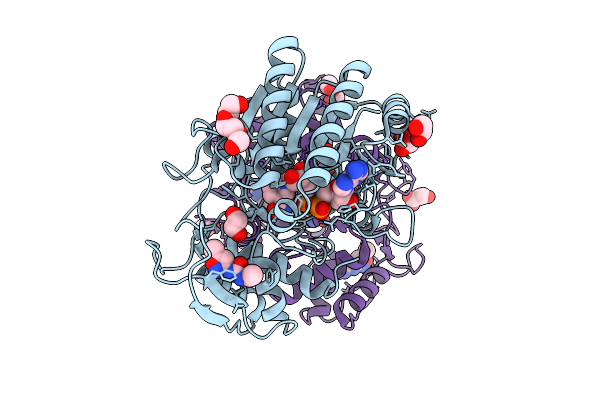 |
Structure Of Udp-Galactose-4-Epimerase (Gale) Bound To Fragment From Diamond Xchem Experiment.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CARBOHYDRATE Ligands: NAD, JGA, MLI, PGE, EDO, CL |
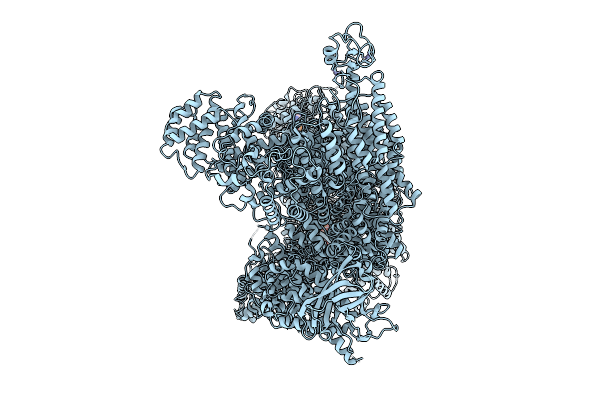 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: LIGASE Ligands: ATP, MG, ZN |
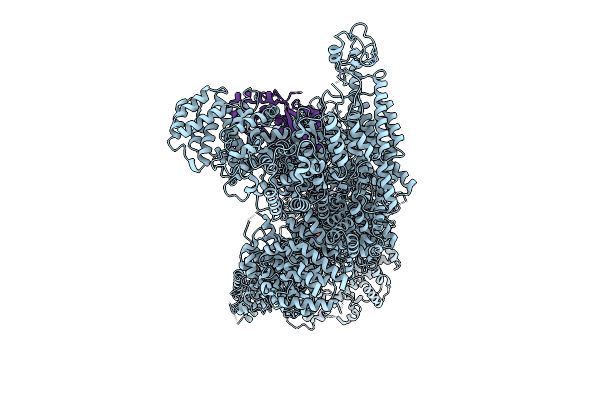 |
Cryo-Em Structure Of Mouse Rnf213:Ube2L3 Transthiolation Intermediate, Chemically Stabilized, And Atpgs
Organism: Mus musculus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: LIGASE Ligands: ATP, MG, ZN, ADP |
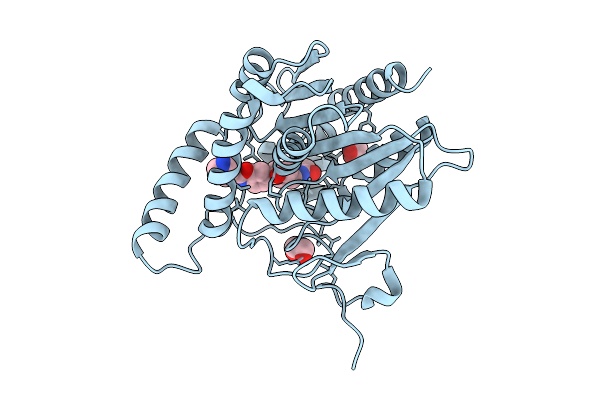 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: A1IZ6, EDO |

