Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
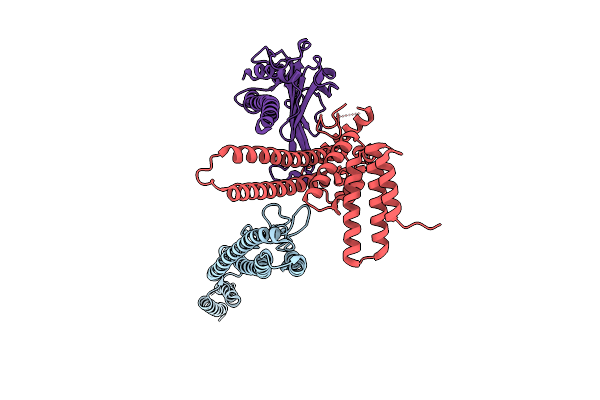 |
Straight And Symmetrical Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
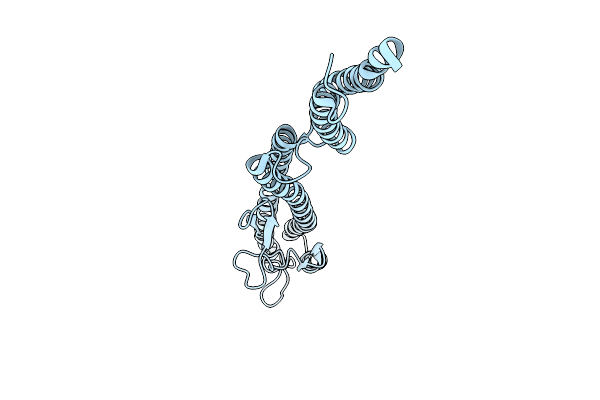 |
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
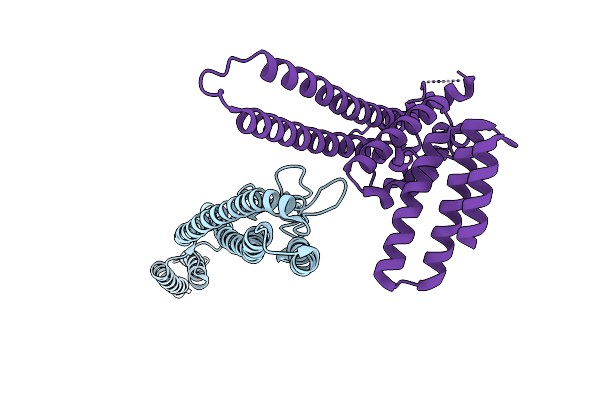 |
Straight And Symmetrical Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa Deleted Fcpb Strain
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
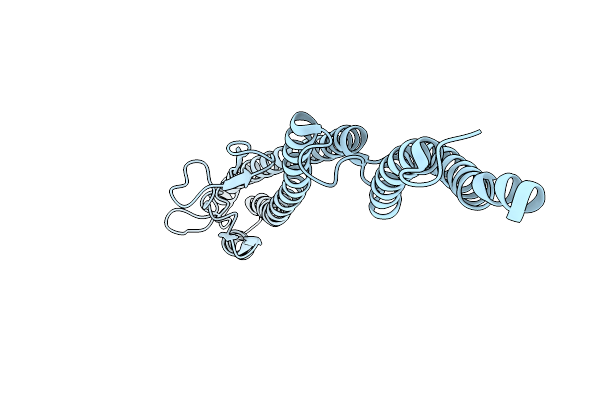 |
Core Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa Deleted Fcpb Strain
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: LMN, ZMA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: LMN, ZMA |
 |
Joint X-Ray/Neutron Room Temperature Structure Of Perdeuterated Leca Lectin In Complex With Deuterated Galactose
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION, NEUTRON DIFFRACTION Release Date: 2025-12-24 Classification: SUGAR BINDING PROTEIN Ligands: GLA, CA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
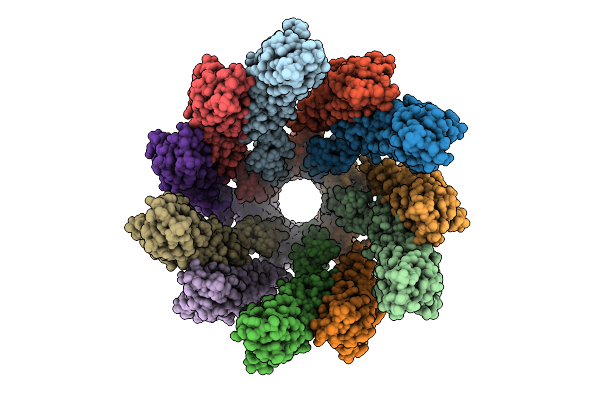
Core Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa Wild Type
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
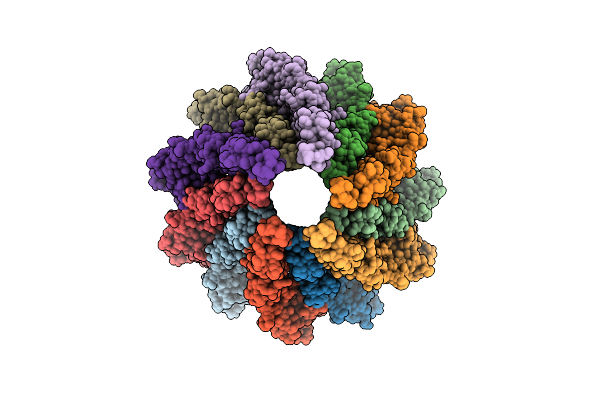
Core Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa From The Flaa2-Complemented Stain
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
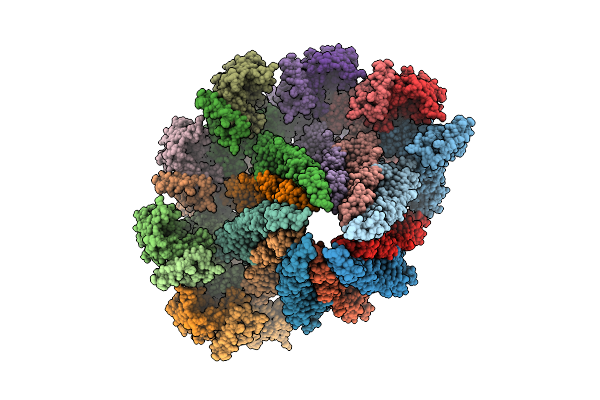
Core Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa From The Deleted Fcpb_Cl13 Strain
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
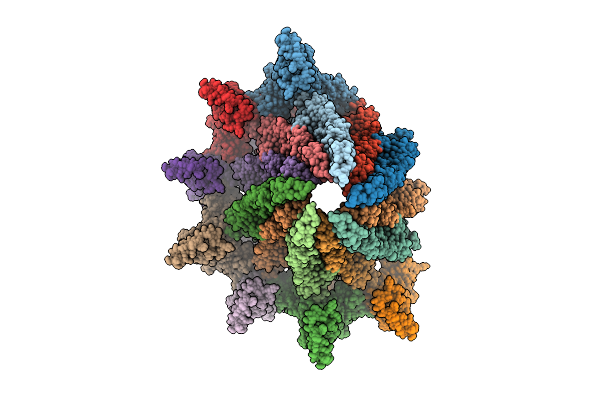
Sheathed Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa From The Flaa2-Complemented Stain
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
Sheathed Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa From The Deleted Fcpb_Cl13 Strain
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
Sheathed Filament Of The Spirochete Periplasmic Flagella Of Leptospira Biflexa Wild Type
Organism: Leptospira biflexa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MOTOR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE/INHIBITOR Ligands: A1C2U, EDO |
 |
Mycobacterium Tuberculosis Relbe1 Toxin-Antitoxin System; Rv1247C (Relb1 Antitoxin), Rv1246C (Rele1 Toxin)
Organism: Mycobacterium tuberculosis h37rv
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TOXIN Ligands: CL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Human Prkcbp1 Zinc Finger Mynd-Type Containing 8 With Pentaglutamate Tag
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: EDO, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: NUCLEAR PROTEIN Ligands: GOL, A1BI5, EDO |