Search Count: 4,169
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: SIGNALING PROTEIN Ligands: SO4 |
 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
 |
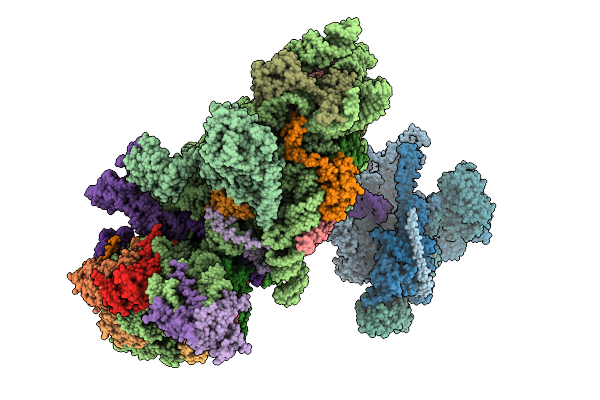
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
 |
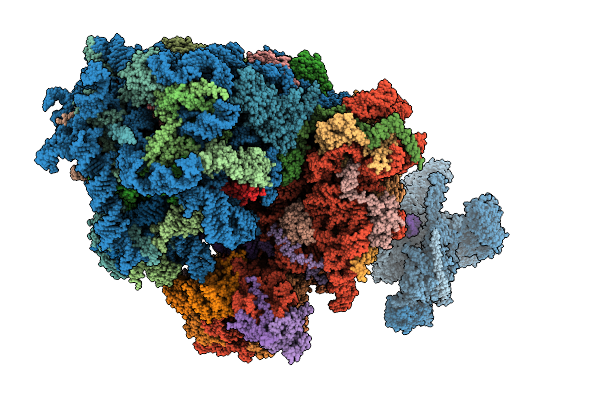
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
 |
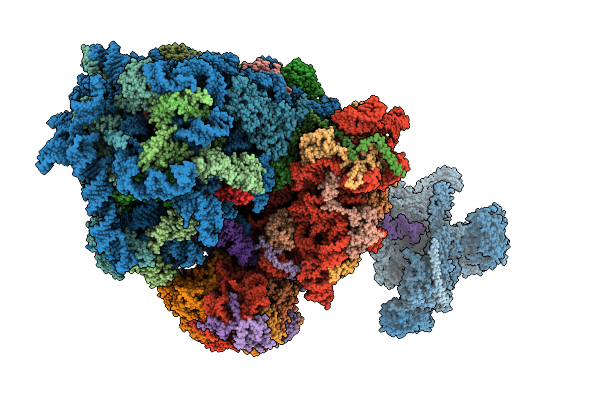
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: CIT, EDO |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: CU, CL, IMD, EDO, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: NI, IMD, GOL, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: ZN, IMD, EDO, GOL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: CO, IMD, GOL, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: DE NOVO PROTEIN Ligands: TB, EDO, GOL, NA |
 |
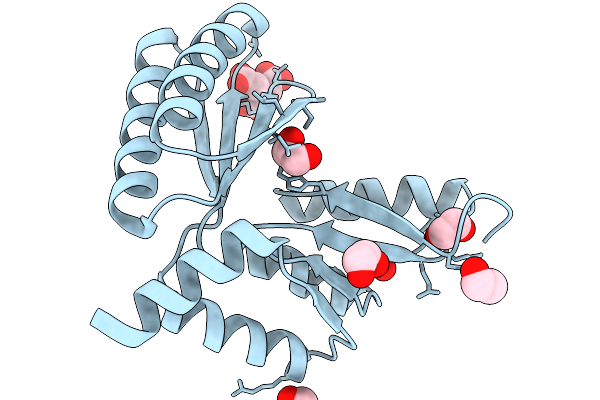
Structure Of Collectin-11 (Cl-11) Carbohydrate-Recognition Domain In Complex With L-Fucose
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: IMMUNE SYSTEM Ligands: CA, FUC, FUL, TRS |
 |
Structure Of Thioferritin (Pfdpsl) With Ferrihydrite Growth At A Single Three-Fold Pore.
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: METAL BINDING PROTEIN Ligands: FE, OXY, O |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, A1BM0 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, PGE, A1BMX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: CA, PGE, EDO, A1BMY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE Ligands: A1CA9 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: LIGASE Ligands: A1CA8 |

