Search Count: 1,04,560
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: PEPTIDE BINDING PROTEIN Ligands: EDO, IMD, PEG |
 |
Sars-Cov-2 Papain-Like-Protease (Plpro) In Complex With Inhibitor Linagliptin
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: 356, GOL |
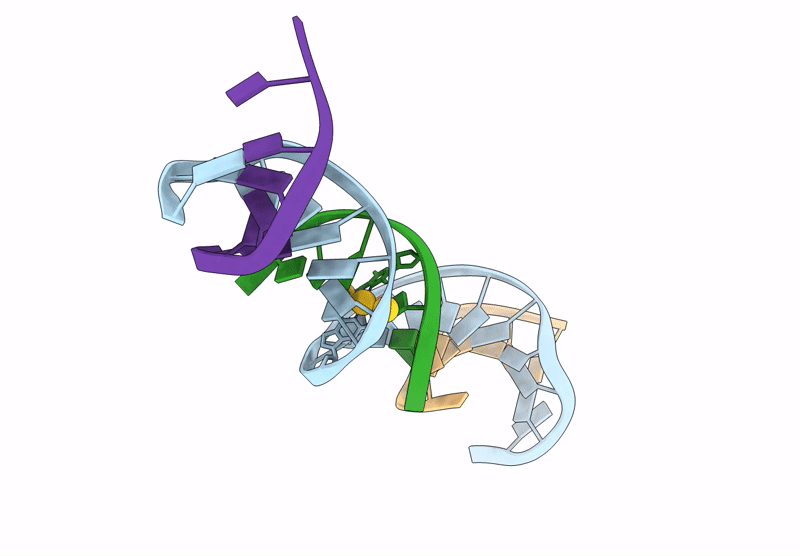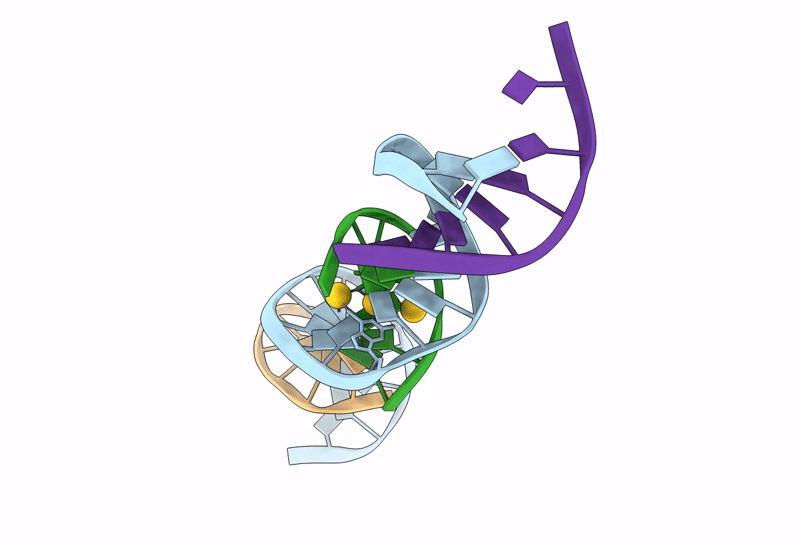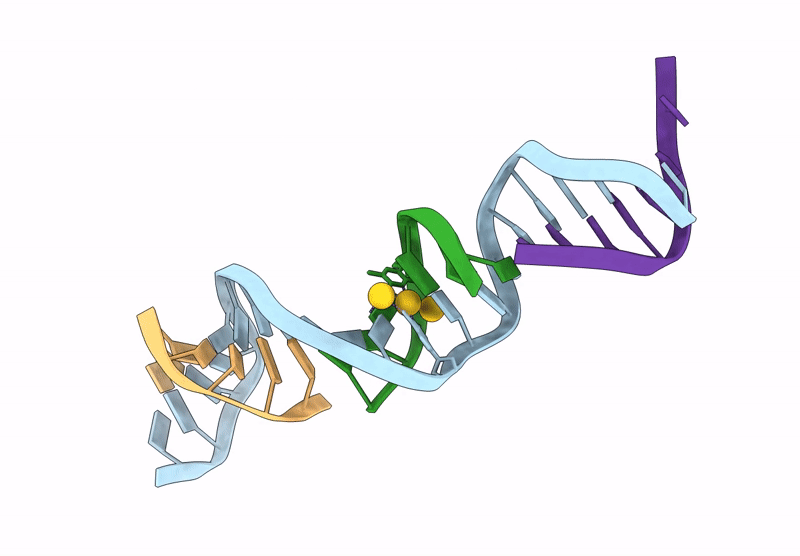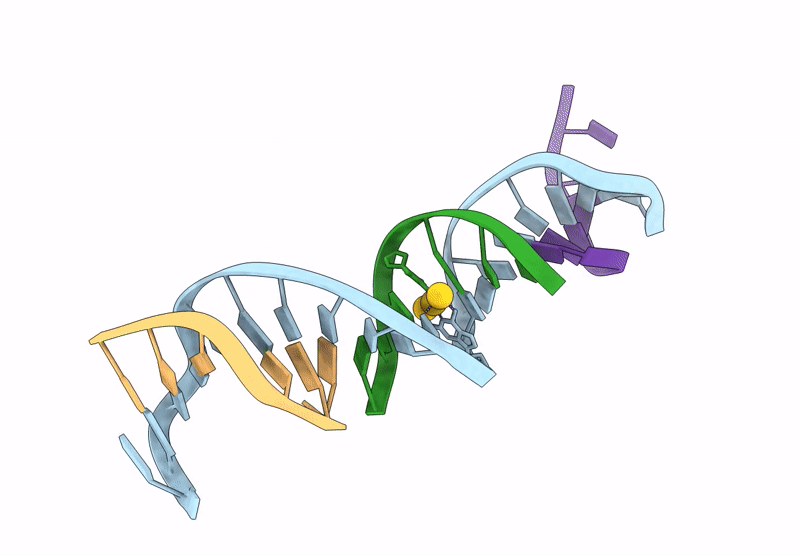 |
[Q:(Au+)3:F_Ph11] Metal-Mediated Dna Base Pair In A Self-Assembling Rhombohedral Lattice
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: DNA Ligands: AU |
 |
Q108K:K40L:T51V:T53C:R58Y:Y19W:Q38L:Q4A:T29L Mutant Of Hcrbpii In Complex With Fr1V In The Dark State At Ph 3.0
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: RETINOL BINDING PROTEIN Ligands: ACT, GOL, ZFJ |
 |
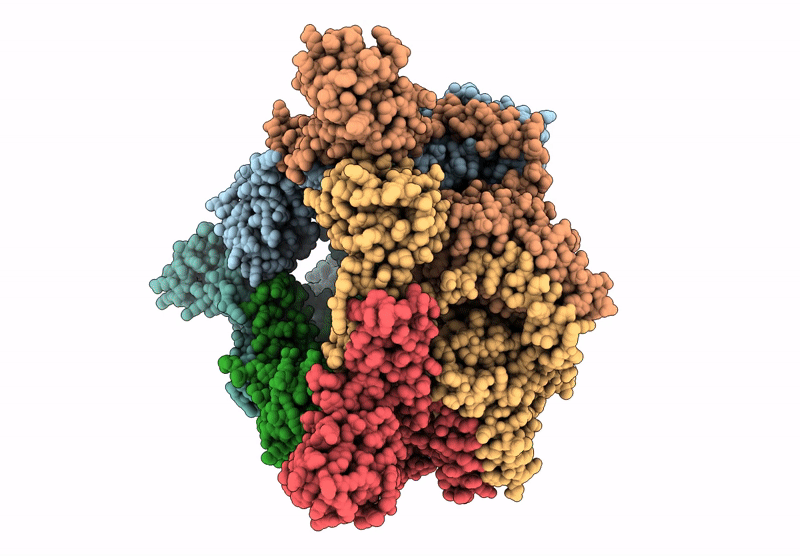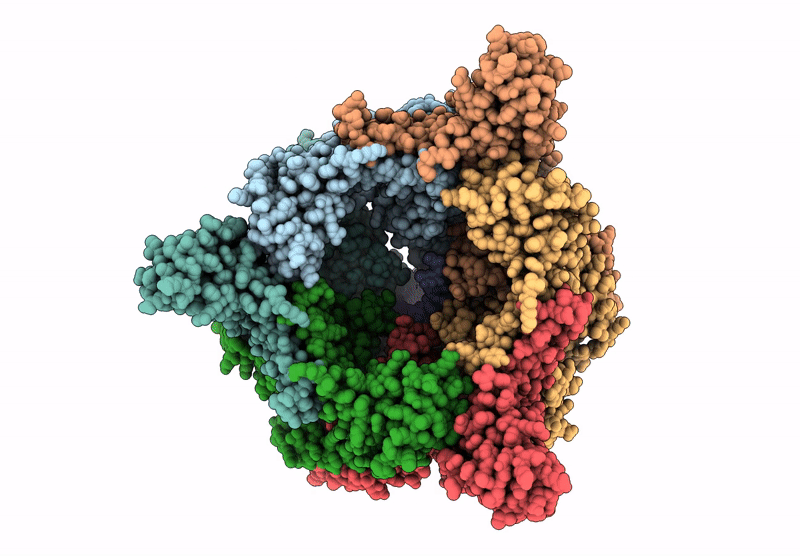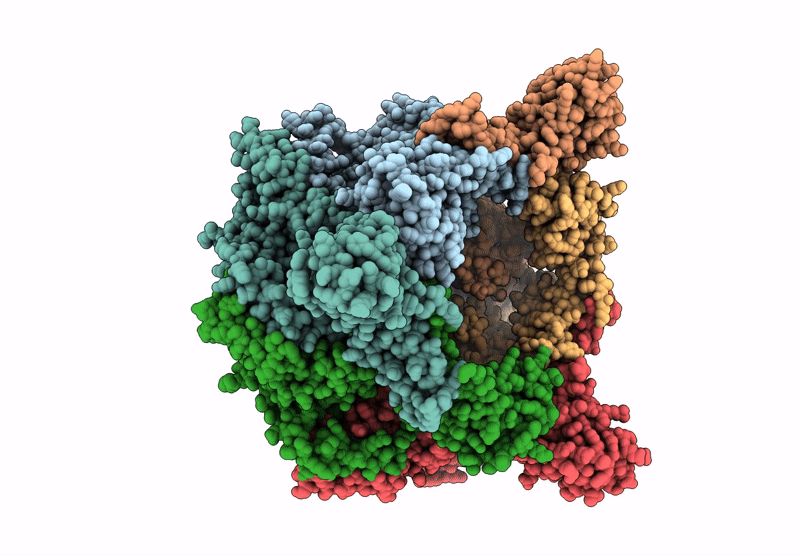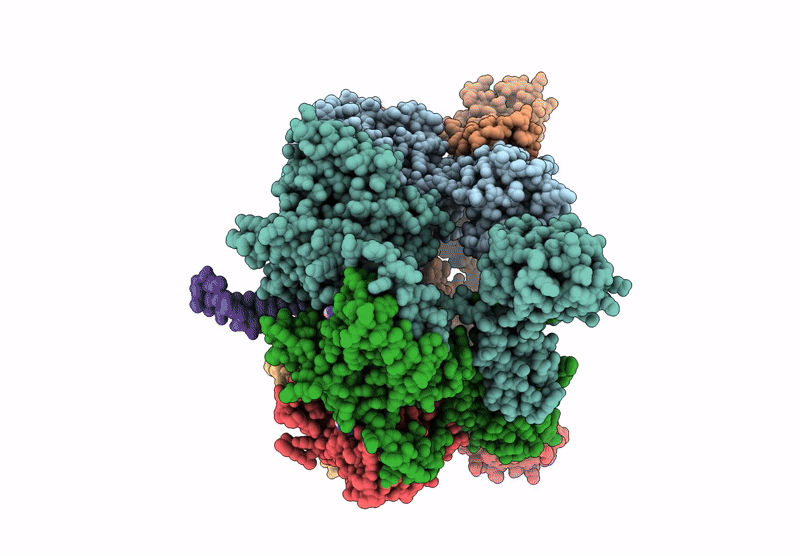
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: LYASE Ligands: ZN, A1A4G |
 |
Organism: Vibrio cholerae, Vibrio cholerae o1
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: DNA BINDING PROTEIN/DNA Ligands: AGS, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSPORT PROTEIN Ligands: T3 |
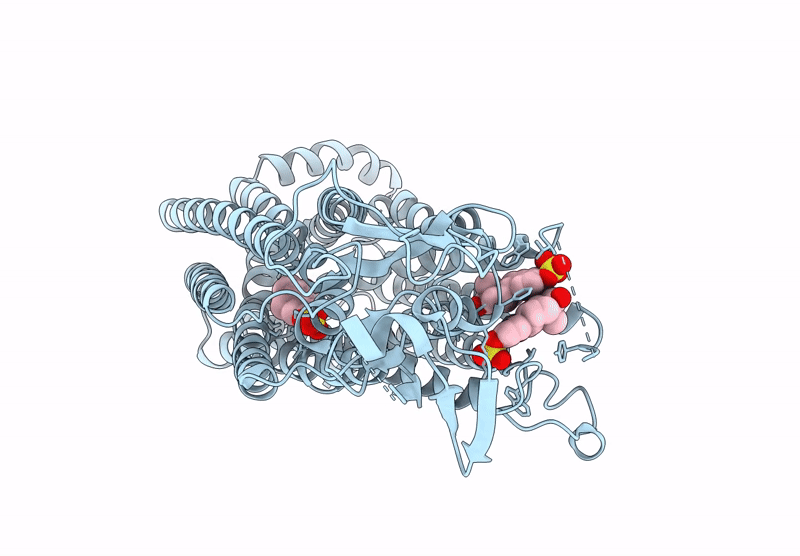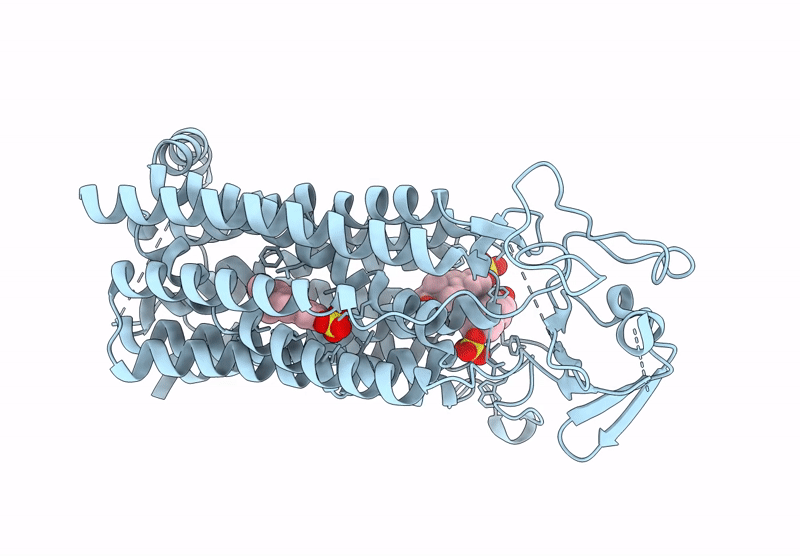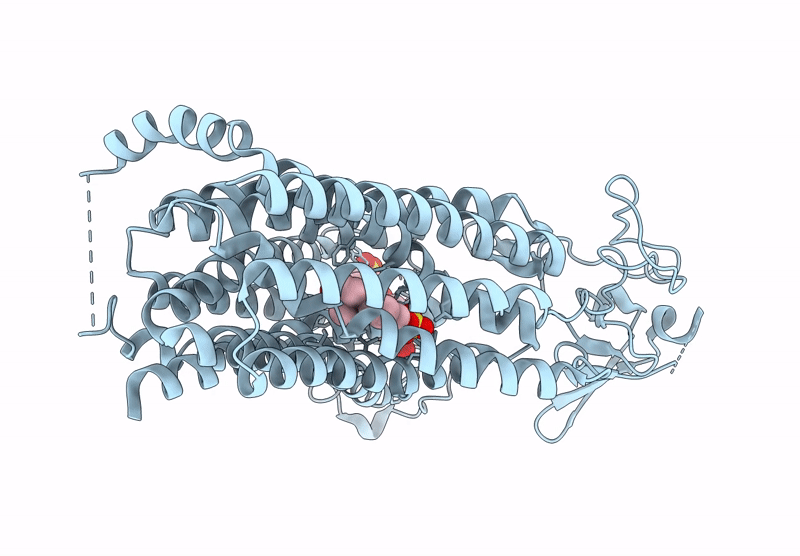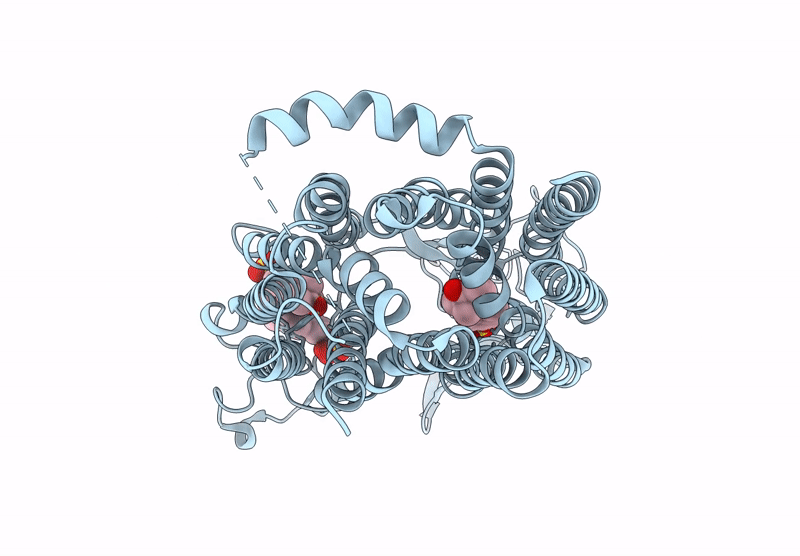 |
Cryo-Em Structure Of Human Oatp1C1 F240A Mutant In Complex With Estrone 3-Sulfate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSPORT PROTEIN Ligands: FY5 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: TRANSPORT PROTEIN Ligands: T44, Y01 |
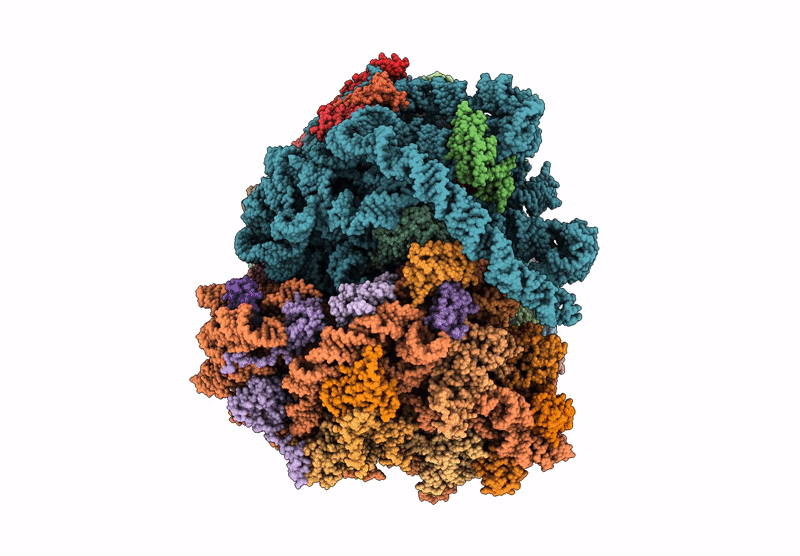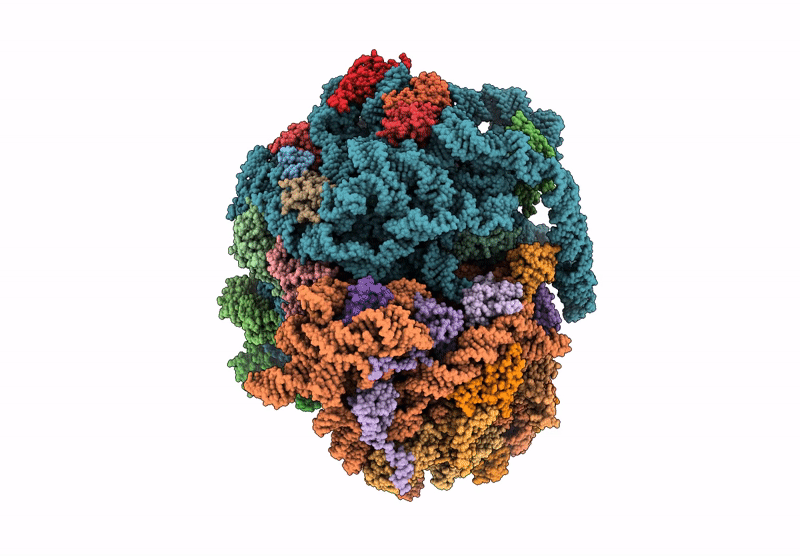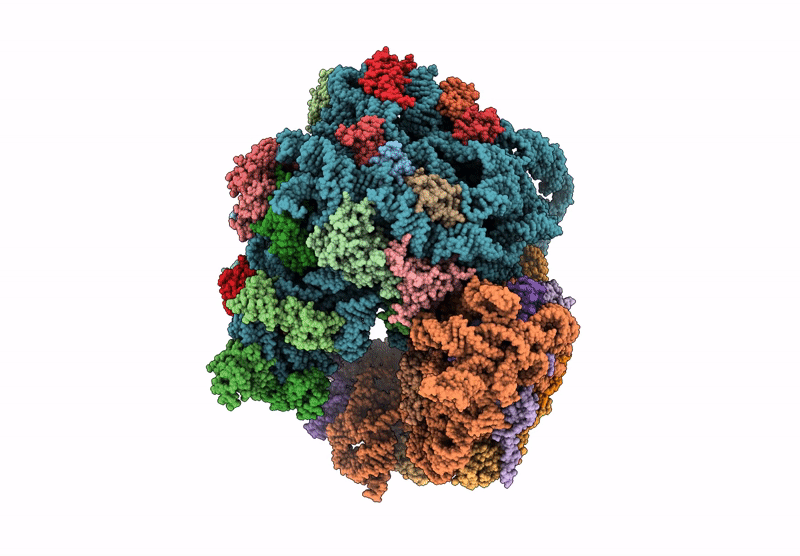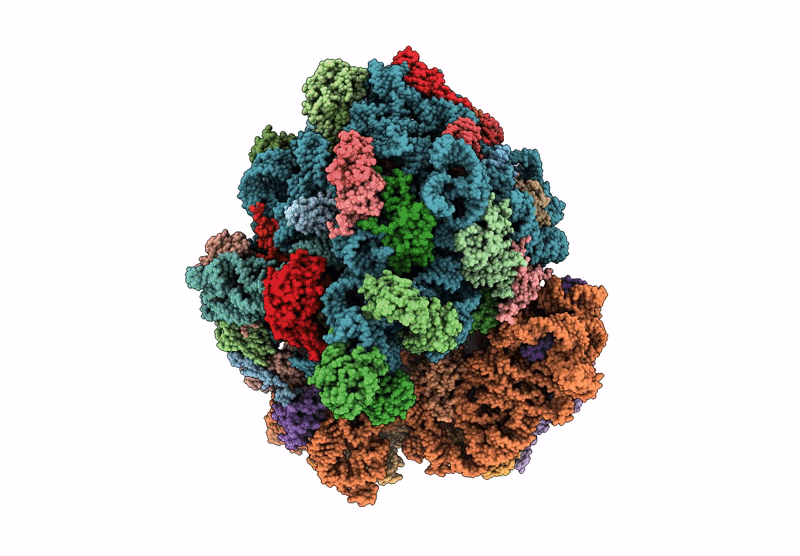 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: RIBOSOME Ligands: MG |
 |
Organism: Mycolicibacterium smegmatis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: RIBOSOME Ligands: MG |
 |
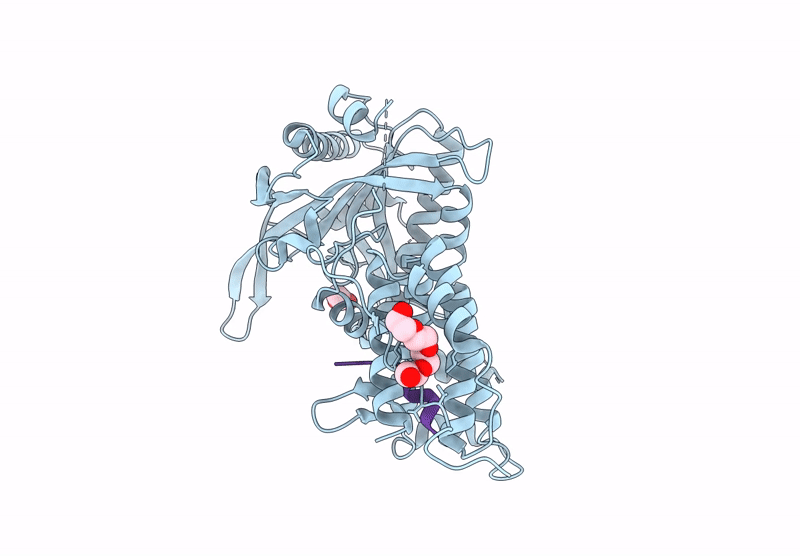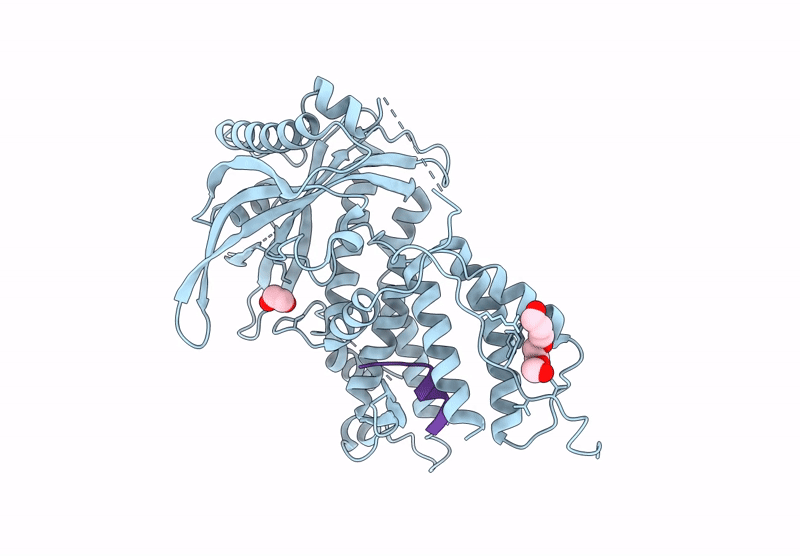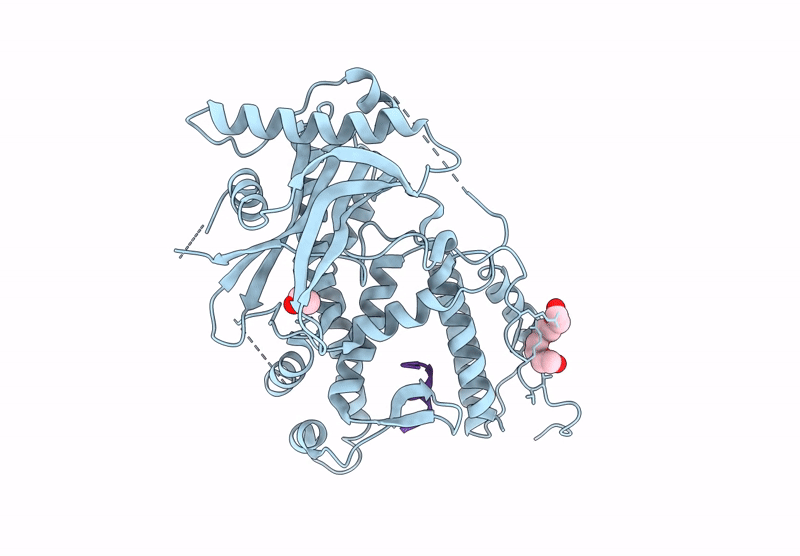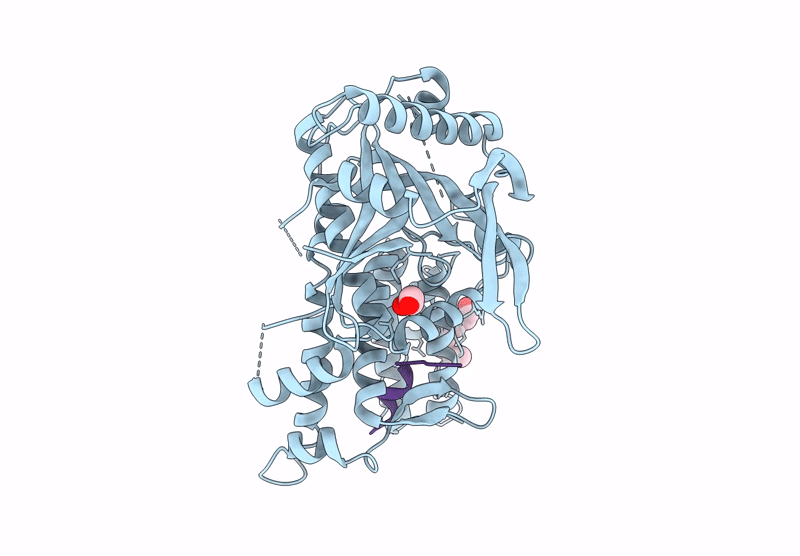
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: CYTOKINE Ligands: NAG, A1H79 |
 |
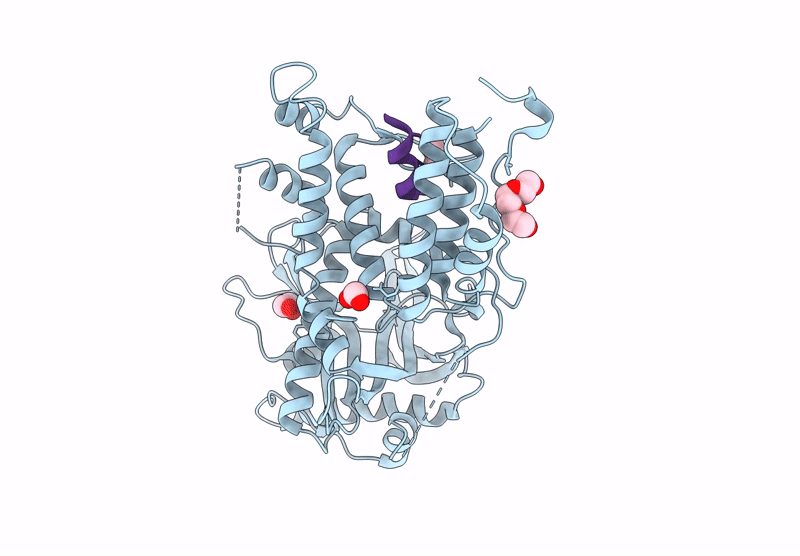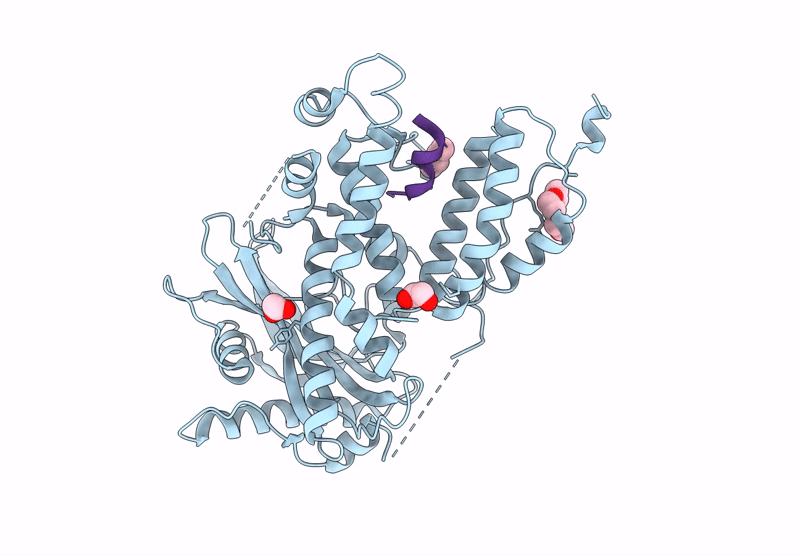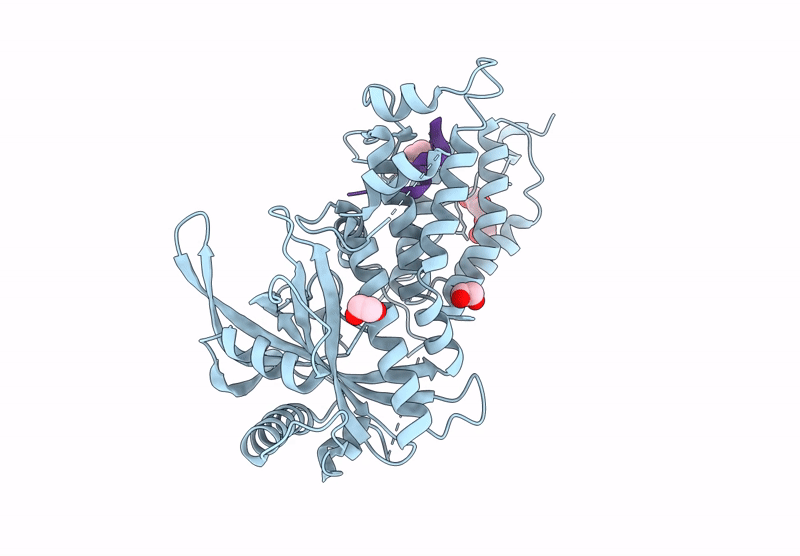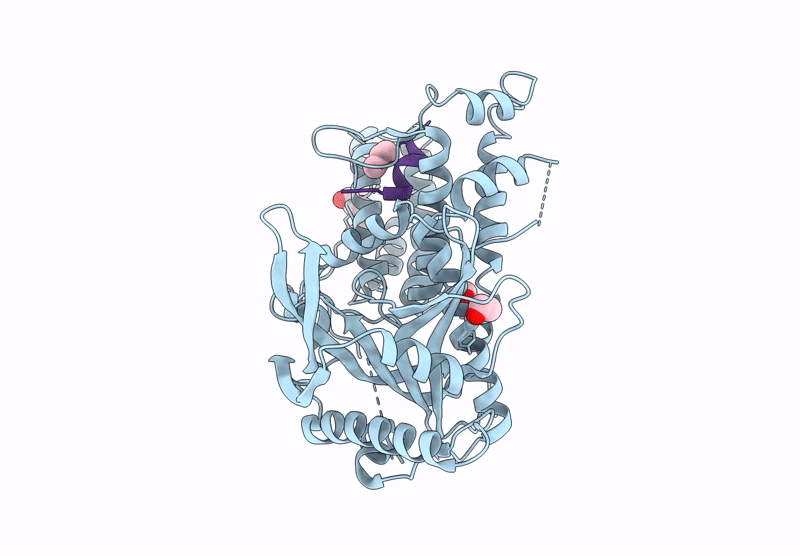
Influenza Polymerase A C-Terminal Domain Of Pa Subunit With Peptide Inhibitor Containing Two Norleucines
Organism: Influenza a virus (a/california/07/2009(h1n1)), Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: EDO, PG4 |
 |
Influenza Polymerase A C-Terminal Domain Of Pa Subunit With Stapled Peptide Inhibitor
Organism: Influenza a virus (a/california/07/2009(h1n1))
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: PGE, EDO, A1IG4 |
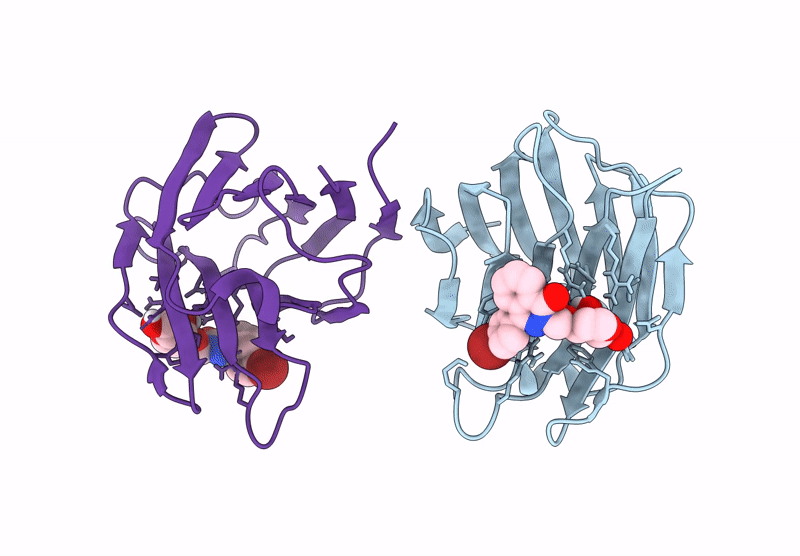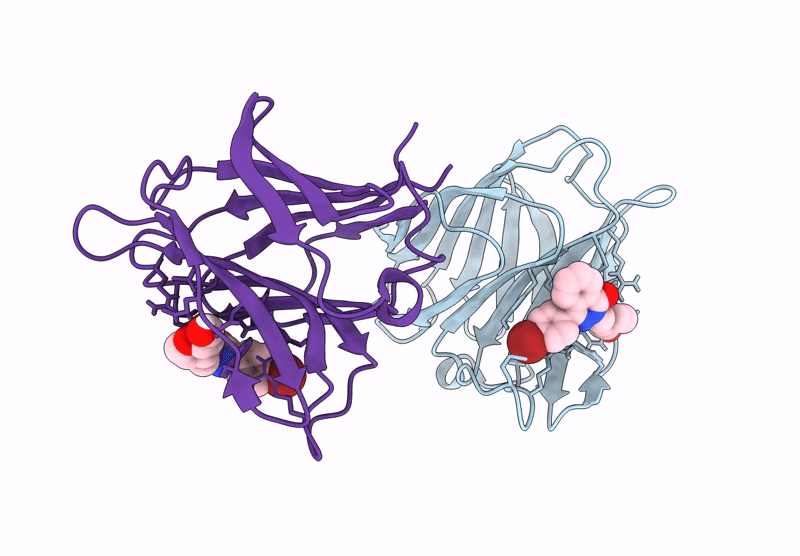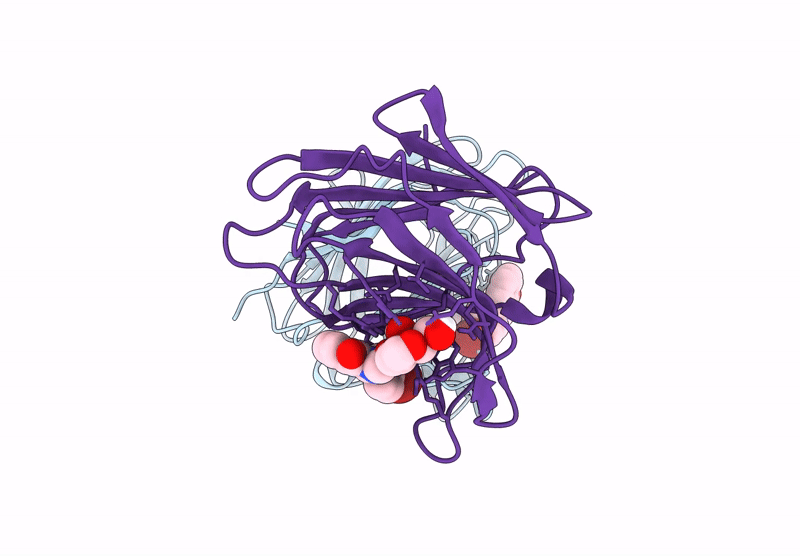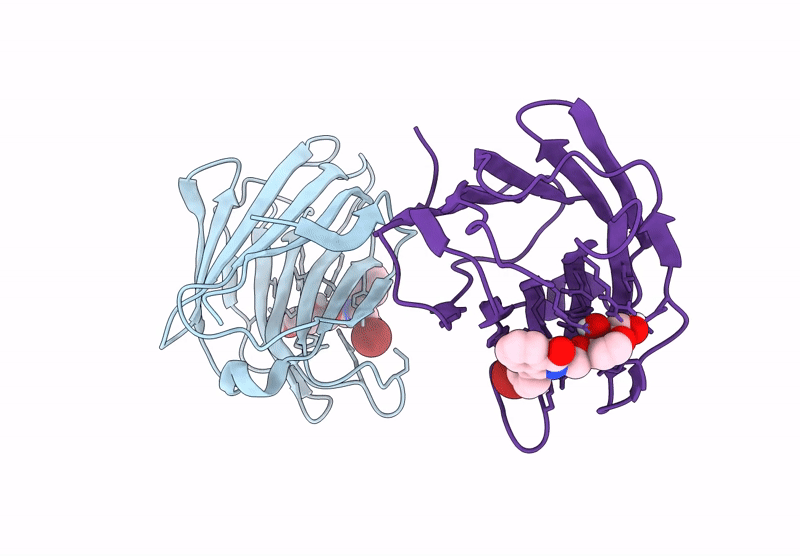 |
Galectin-8 N-Terminal Carbohydrate Recognition Domain In Complex With 4-(Bromophenyl)Phthalazinone D-Galactal Ligand
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: IMMUNE SYSTEM Ligands: V19, CL |
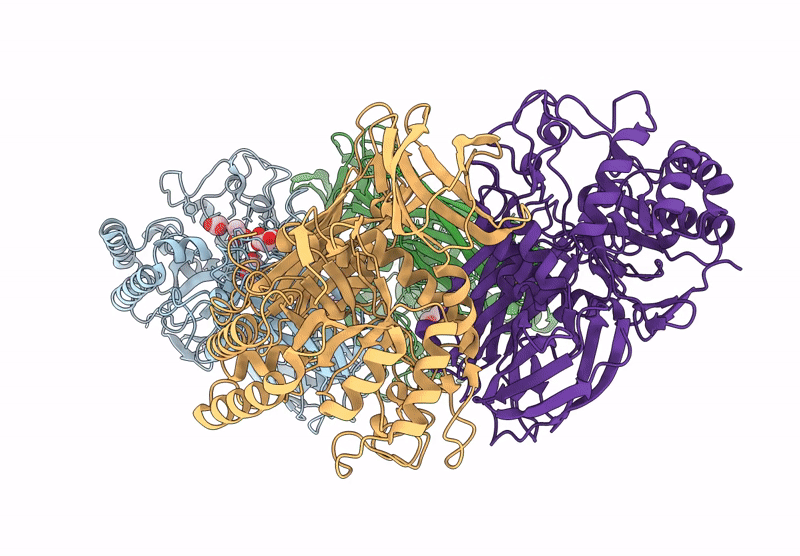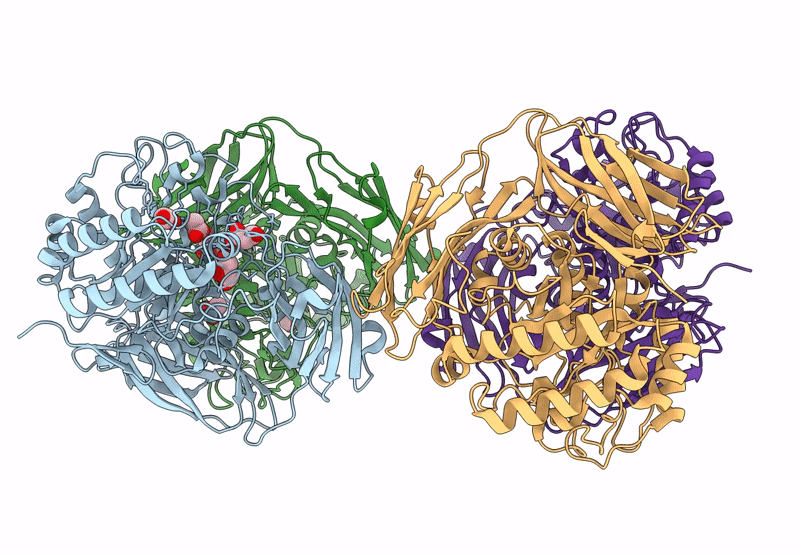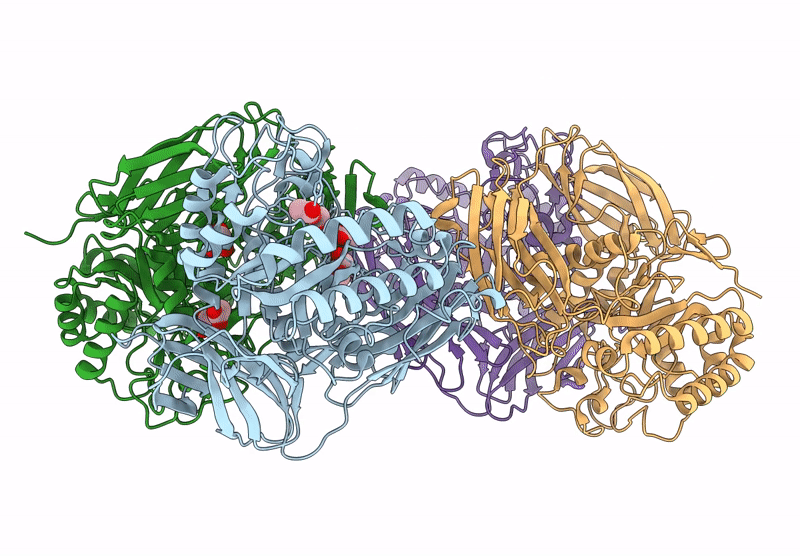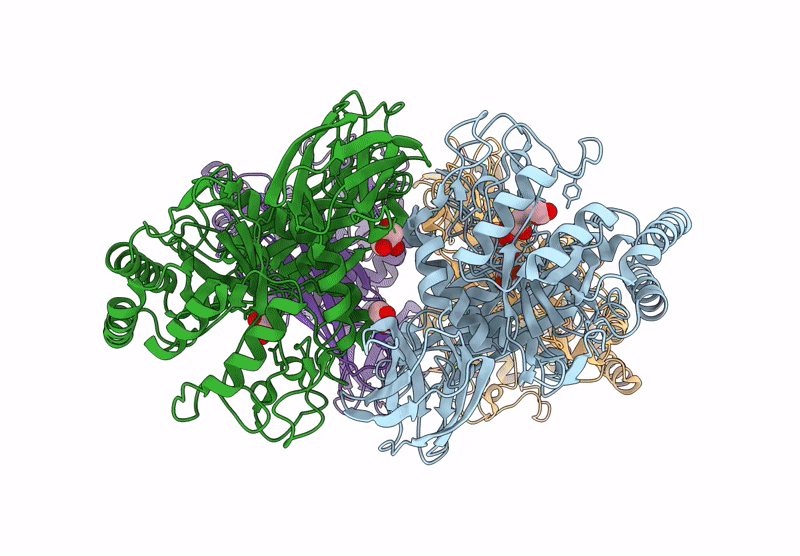 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum, Wild Type Semet Derivative (Lagh157)
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: ACT, GOL, MLA |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fructose-6-Phosphate
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: F6P, CL |
 |
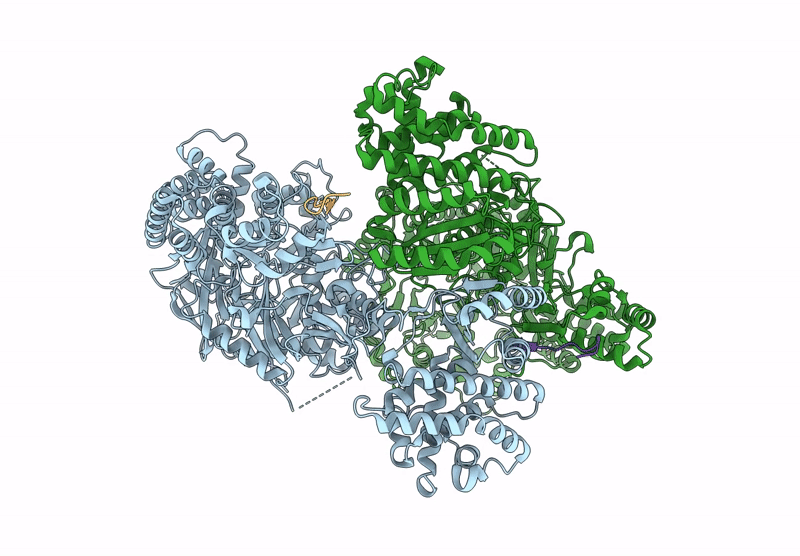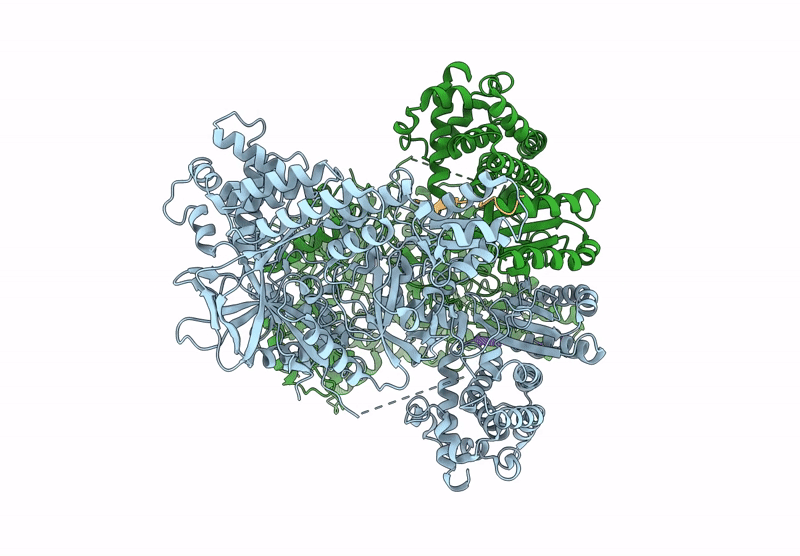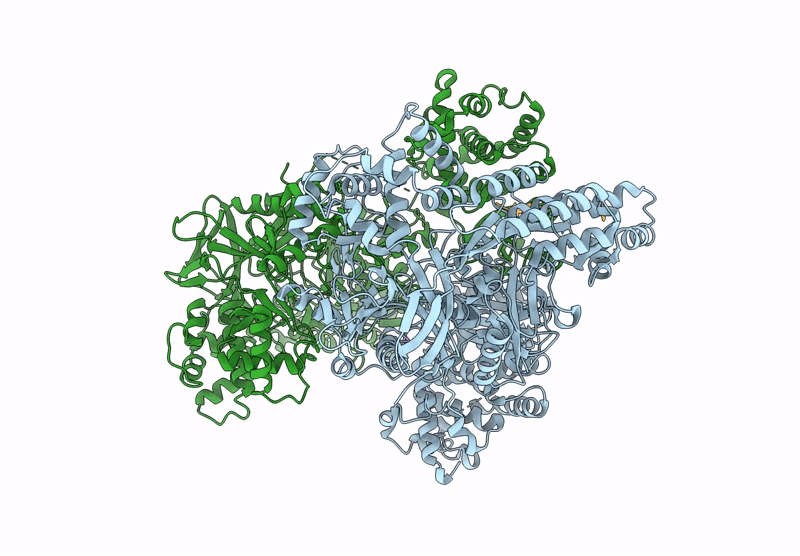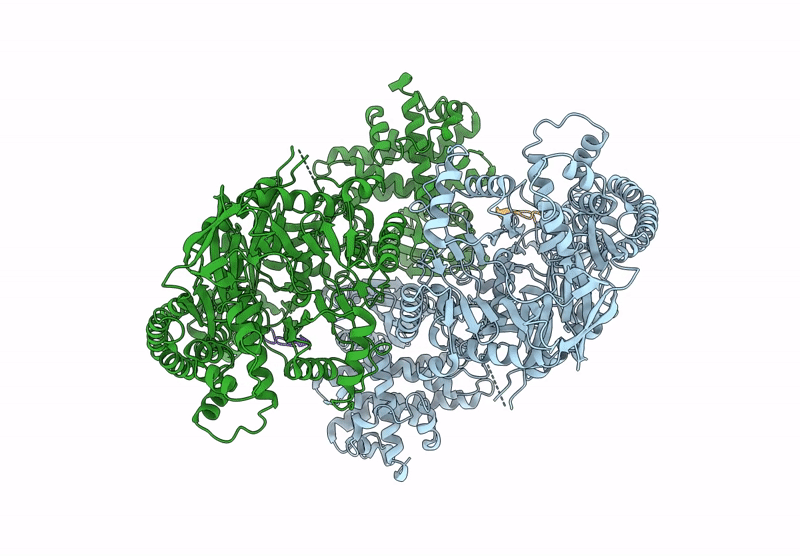
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley In The Apo State
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Madb, A Class I Dehydrates From Clostridium Maddingley, In Complex With Its Substrate
Organism: Clostridium sp. maddingley mbc34-26
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN |

