Search Count: 46,743
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fructose-6-Phosphate
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: F6P, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment
Organism: Candida albicans
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: A1IHU, PA5 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: MN, NAG, ACY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: NAG, MN, UD2, ACY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: UDP, MN, ACY |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: A1IUN, EDO |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: VIRAL PROTEIN Ligands: A1IUM, EDO |
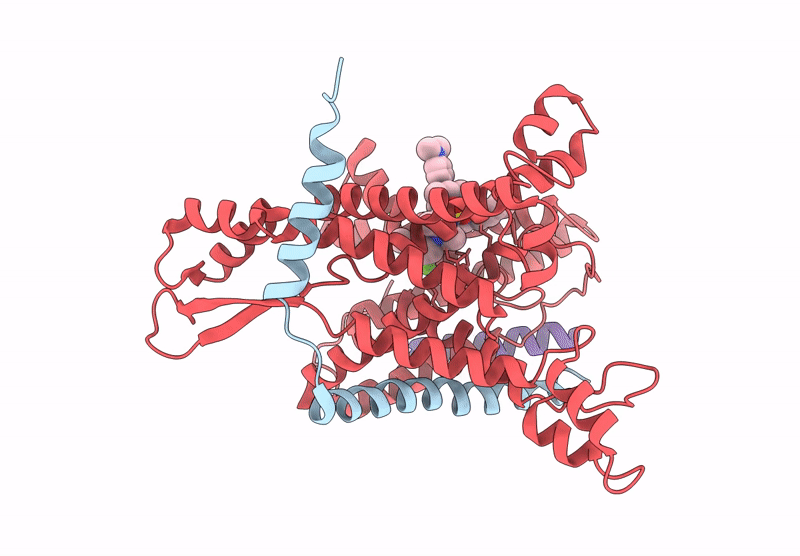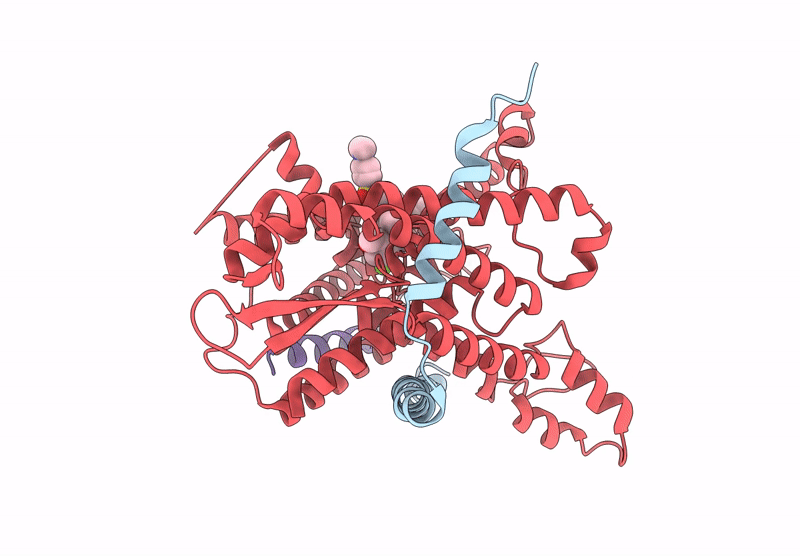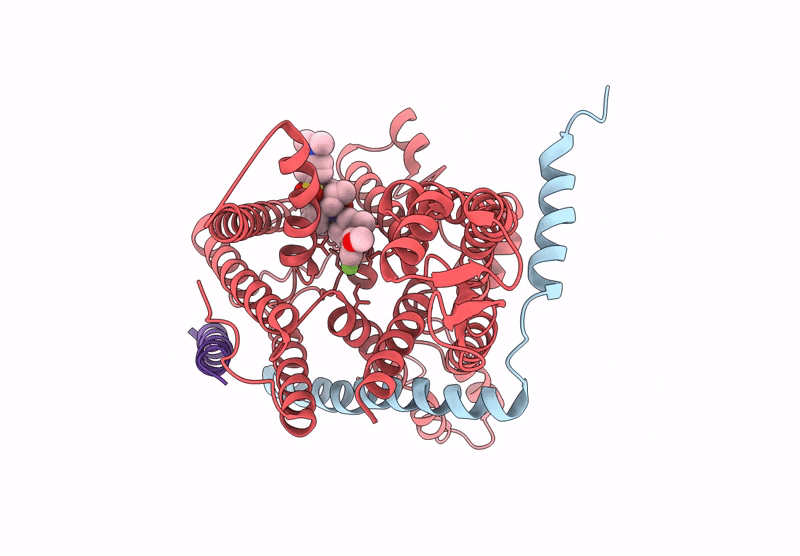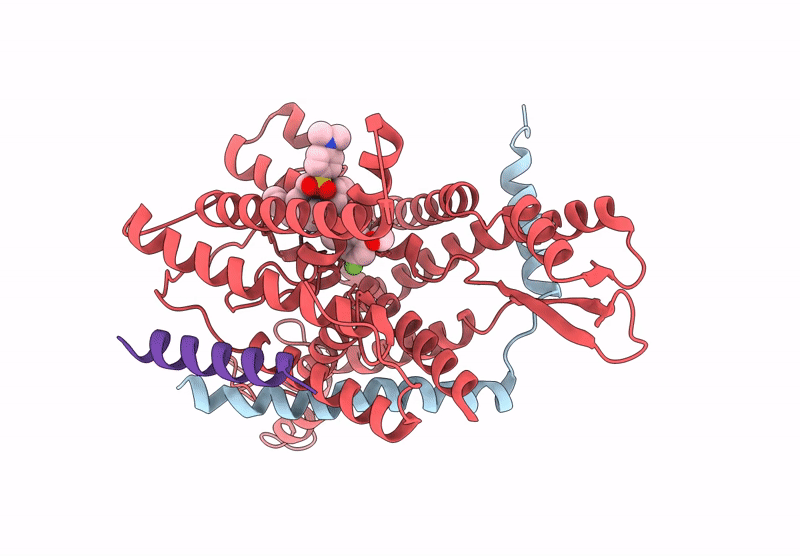 |
Pre-Clinical Characterization Of Novel Multi-Client Inhibitors Of Sec61 With Broad Anti-Tumor Activity
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: PROTEIN TRANSPORT Ligands: A1A2B |
 |
Room Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 95% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Room-Temperature Structure Of Kr2 Rhodopsin In Pentameric Form At 85% Relative Humidity
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: MEMBRANE PROTEIN Ligands: RET, NA, OLC, LFA |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 95% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Collected At Euxfel Spb/Sfx With Hve Injection Method
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 95% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Hen Egg-White Lysozyme Structure Embedded In Lcp Medium At 85% Relative Humidity
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE |
 |
Crystal Structure Of The Peptidyl-Prolyl Isomerase (Ppiase) From E. Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: ISOMERASE Ligands: GOL, CD |
 |
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Zea Mays 3-Phosphoglycerate Dehydrogenase S282L Mutant
Organism: Zea mays
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: OXIDOREDUCTASE |

