Search Count: 9,873
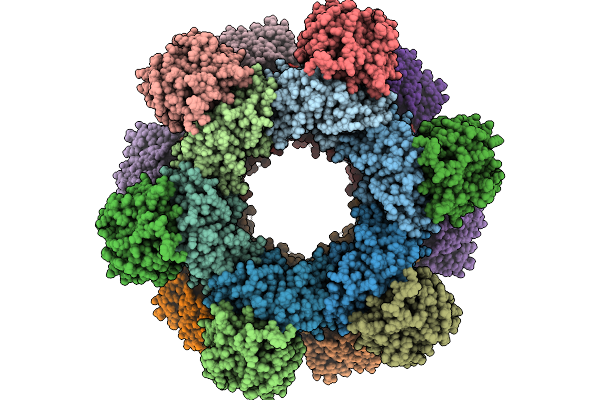 |
Cryo-Em Structure Of A Designed Pyridoxal Phosphate (Plp) Synthase Fused To A Designed Circumsporozoite Protein Antigen From Plasmodium Falciparum (Csp-P1-Csp And Csp-P2-Csp)
Organism: Plasmodium falciparum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: BIOSYNTHETIC PROTEIN |
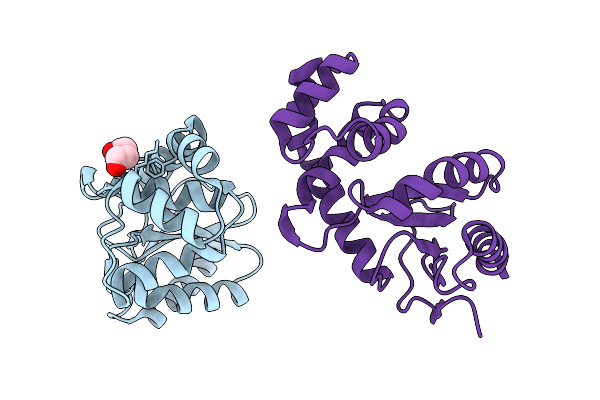 |
Organism: Bordetella pertussis
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: OXIDOREDUCTASE Ligands: PEG |
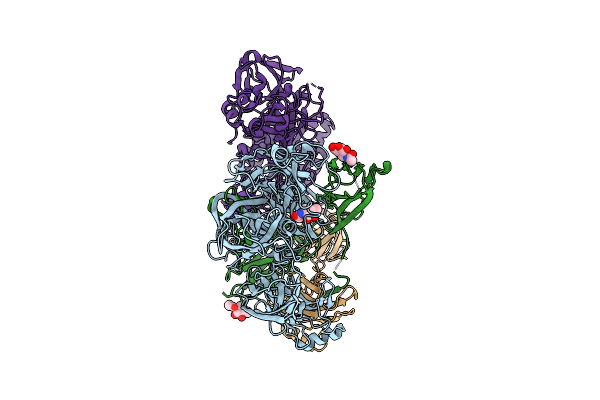 |
Structure Of The Complement Classical And Lectin Pathway Proconvertase, C4B2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: NAG, MG |
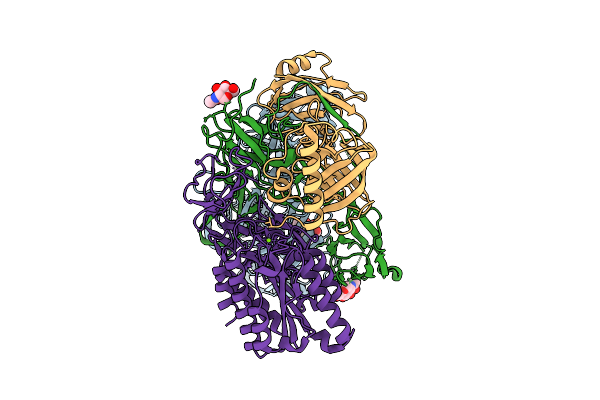 |
Structure Of The Complement Classical And Lectin Pathway Proconvertase, C4B2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: NAG, MG |
 |
Structure Of The Complement Classical And Lectin Pathway C3 Convertase In Complex With Substrate C3
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: NAG, MG |
 |
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: NAG, MG |
 |
X-Ray Structure Of Furin (Pcsk3) In Complex With The Biphenyl-Derived Compound 26 (Mi2456)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1JA6, CA, NA, CL, DMS, NAG |
 |
X-Ray Structure Of Furin (Pcsk3) In Complex With The Biphenyl-Derived Compound 42 (Mi2464)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: CA, NA, CL, DMS, NAG, A1JA1 |
 |
X-Ray Structure Of Furin (Pcsk3) In Complex With The Biphenyl-Derived Compound 34 (Mi2470)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: CA, NA, CL, DMS, NAG, A1JA2 |
 |
X-Ray Structure Of Furin (Pcsk3) In Complex With The Biphenyl-Derived Compound 27 (Mi2471)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1JA3, CA, NA, CL, DMS, NAG |
 |
X-Ray Structure Of Furin (Pcsk3) In Complex With The Biphenyl-Derived Compound 13 (Mi3102)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: CA, NA, CL, DMS, NAG, A1JA5 |
 |
X-Ray Structure Of Furin (Pcsk3) In Complex With The Biphenyl-Derived Compound 24 (Mi3140)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1JA4, CA, NA, CL, DMS, NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1JH5 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1JKT, EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: A1JKZ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: HYDROLASE Ligands: CL, A1JK6 |
 |
Group Ii Intron Assembly Intermediate Domain 1, 2, 3 And 4 "Fully Open" State
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE Ligands: MN |

