Search Count: 8,077
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
 |
Group Ii Intron Assembly Intermediate Domain 1, 2, 3 And 4 "Partly Open" State
Organism: Oceanobacillus iheyensis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: RNA |
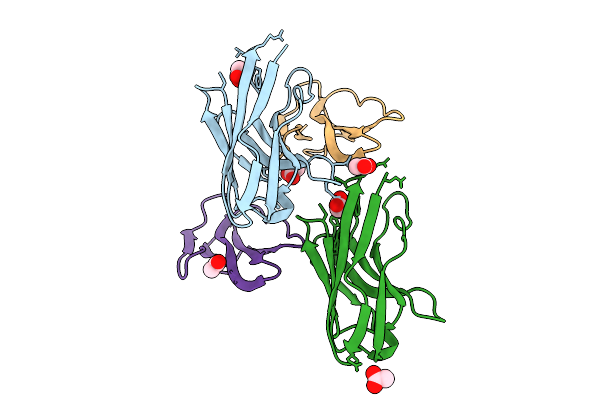 |
Co-Crystal Of Broadly Neutralizing Biparatopic Vhh In Complex With Cardiotoxin (P01468) Naja Pallida
Organism: Vicugna pacos, Naja pallida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN Ligands: ACT |
 |
Co-Crystal Of Broadly Neutralizing Vhh In Complex With Short Neurotoxin 1 (P01426) Naja Pallida
Organism: Vicugna pacos, Naja pallida
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TOXIN Ligands: NI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: RNA BINDING PROTEIN Ligands: GOL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: EDO, MES |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: MG, CL |
 |
Organism: Pseudomonas fluorescens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: LYASE Ligands: CL |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TRANSFERASE |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: TOXIN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Release Date: 2025-10-29 Classification: HYDROLASE Ligands: SO4 |
 |
Ternary Protac-Mediated Complex Consisting Of Cereblon, Ddb1 And Brd4-Bd1, Non-Covalently Linked By Jq1-Acn
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: LIGASE Ligands: ZN, A1JM3 |
 |
Organism: Schizosaccharomyces pombe, Murine hepatitis virus strain a59
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: PROTEIN TRANSPORT Ligands: EDO, GOL |
 |
Cryoem Structure Of Mammalian Aap In Complex With Acetyl-Alanyl-Chloromethylketone
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: A1INA |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: A1ING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: ONCOPROTEIN Ligands: ZN, A1IWG |
 |
Cryo-Em Structure Of Acylaminoacyl Peptidase (Aap) In Covalent Complex With Inhibitor Aebsf
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: HYDROLASE Ligands: AES |

