Search Count: 14,767
 |
Organism: Gallus gallus
Method: SOLUTION NMR Release Date: 2025-12-31 Classification: STRUCTURAL PROTEIN |
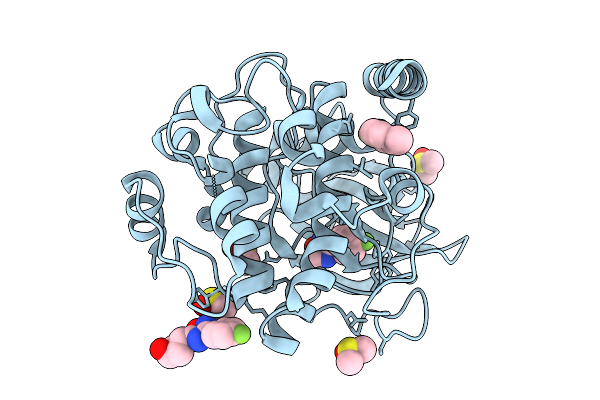 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F001
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1JE8, DMS |
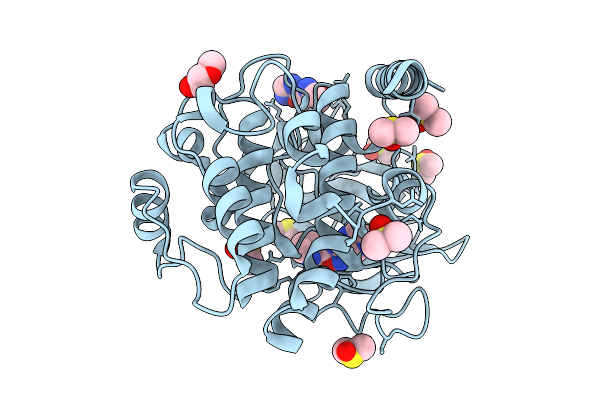 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F005
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PG4, DMS, A1JE7 |
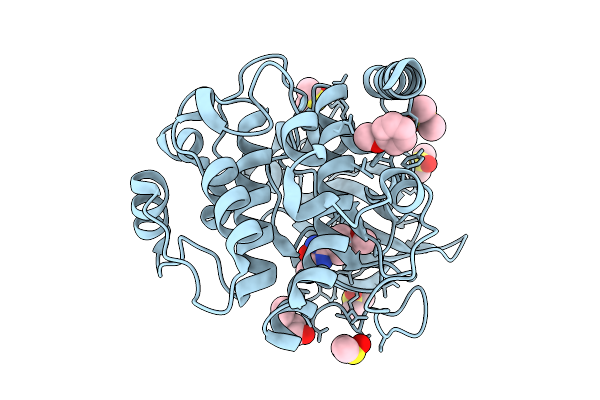 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F009
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE6, PEG |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F012
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE5 |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F024
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE4 |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F030
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PEG, A1JE9, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F032
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE3 |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F055
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PEG, A1JE2, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F058
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, PEG, A1JE1 |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F070
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JE0 |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F073
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PG4, A1JEZ, PEG, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F074
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1JEY |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F102
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JEX |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F134
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: D6V, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F138
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1JEQ, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F145
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: DMS, A1JER |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F168
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1JES, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F184
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: A1JET, DMS |
 |
Crystal Structure Of The Protein-Kinase A Catalytic Subunit From Cricetulus Griseus In Complex With F186
Organism: Cricetulus griseus
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: TRANSFERASE Ligands: PGF, A1JEU, DMS |

