Search Count: 26,439
 |
Structure Of The Proline-Rich Binding Domain Of Tesup-1 In Complex With Dzfc3H1 Peptide
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-02-04 Classification: PROTEIN BINDING Ligands: TLA, ACT |
 |
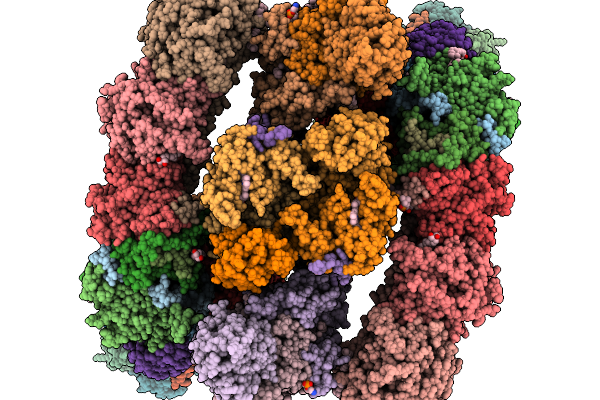
Organism: Paracoccus denitrificans pd1222
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: ELECTRON TRANSPORT Ligands: SF4, NA, FES, FMN, DU0, U10, ZN, CDL, HEM, CA, HEC, HEA, CU, MN, CUA |
 |
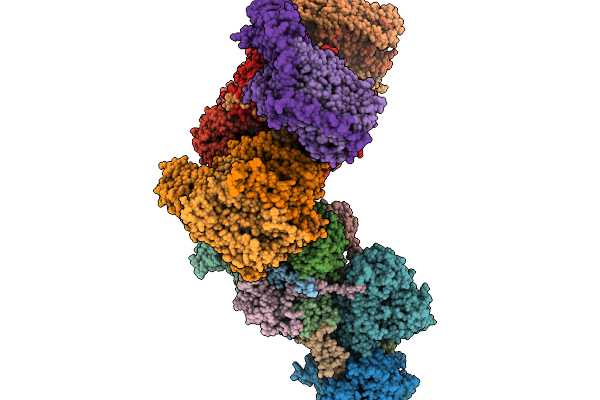
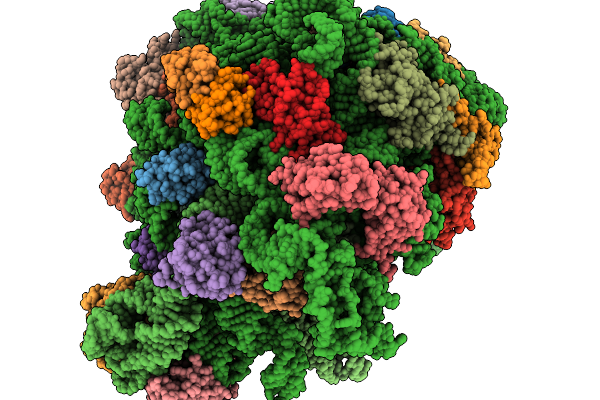
Respiratory Supercomplex Ci2-Ciii2-Civ2 (Megacomplex) From Alphaproteobacterium
|
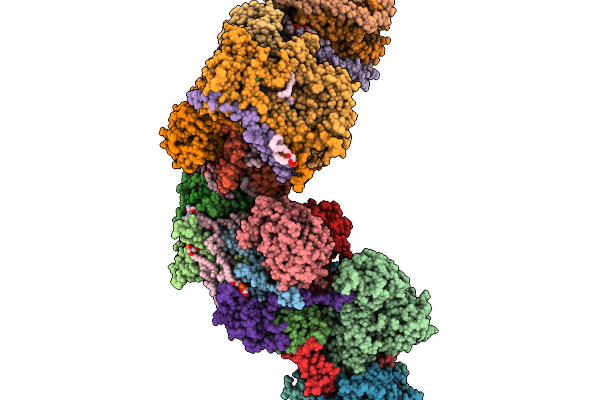 |
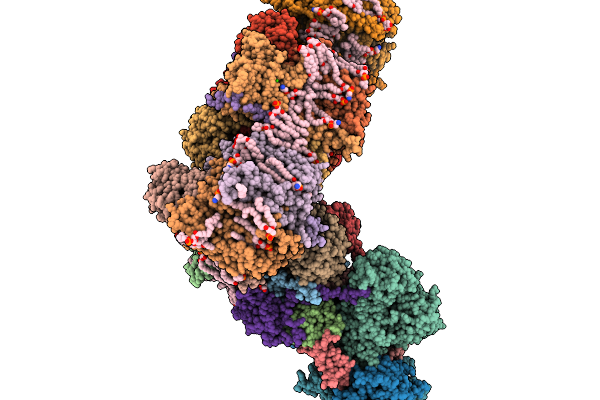
Respiratory Supercomplex Ci1-Ciii2-Civ1 (Respirasome) From Alphaproteobacterium
|
 |
 |
Structure Of E.Coli Ribosome With Filamin Mutant Y719E Nascent Chain At Linker Length Of 47 Amino Acids, With Trna
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RIBOSOME |
 |
Photosynthetic A10B10 Glyceraldehyde-3-Phospahte Dehydrogenase From Spinacia Oleracea.
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Resolution:7.20 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Escherichia coli, Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: SIGNALING PROTEIN Ligands: A1L7J |
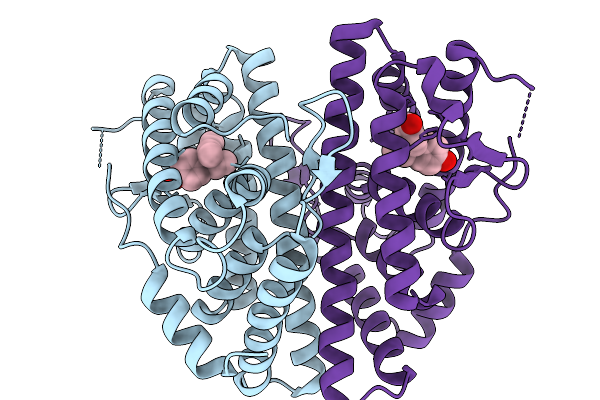 |
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Bisphenol B
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: H3W |
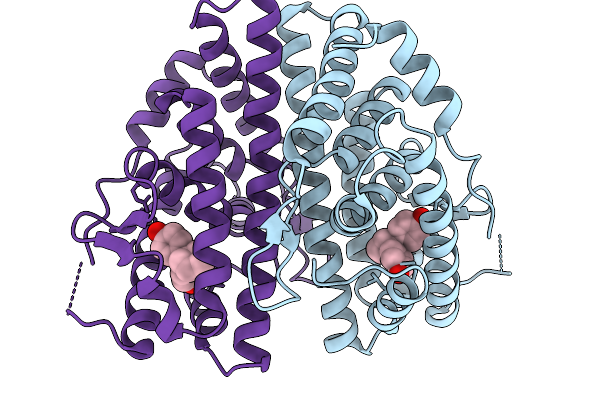 |
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Bisphenol Z
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: BPZ |
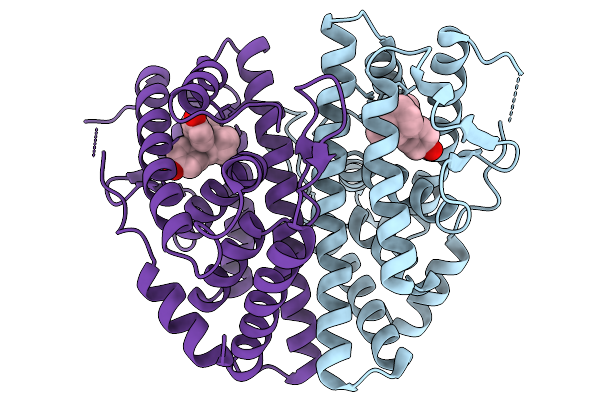 |
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Bisphenol Tmc
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: A1BVK, A1BWR |
 |
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Estradiol And D22 13Mer
Organism: Melanotaenia fluviatilis, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: EST |
 |
Crystal Structure Of Melanotaenia Fluviatilis Estrogen Receptor Alpha Ligand Binding Domain Complexed With Estradiol
Organism: Melanotaenia fluviatilis
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2026-02-04 Classification: NUCLEAR PROTEIN Ligands: EST |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2026-02-04 Classification: IMMUNE SYSTEM Ligands: EDO, CL, PO4 |
 |
Crystal Structure Of Hp1Gamma Chromoshadow Domain In Complex With Kap1 Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2026-02-04 Classification: TRANSCRIPTION |
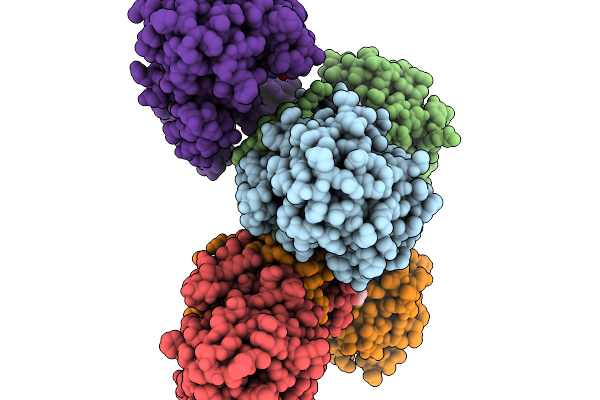 |
Structure Of Pycr1 Complexed With The Allosteric Inhibitor 1-Benzyl-1H-1,3-Benzodiazole-2-Sulfonic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: A1CFK, SO4 |
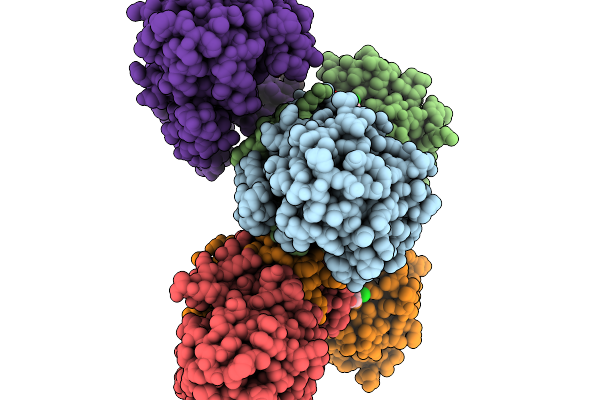 |
Structure Of Pycr1 Complexed With The Allosteric Inhibitor 2-[(2,6-Dichlorophenyl)Amino]Pyridine-3-Sulfonic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE Ligands: A1CFJ, SO4, PEG, PGE |
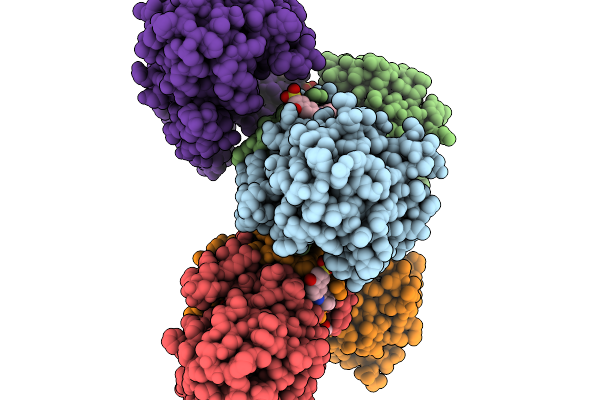 |
Structure Of Pycr1 Complexed With The Allosteric Inhibitor 4-Hydroxy-7-(Phenylamino)Naphthalene-2-Sulfonic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: A1CFI, SO4, PGE |
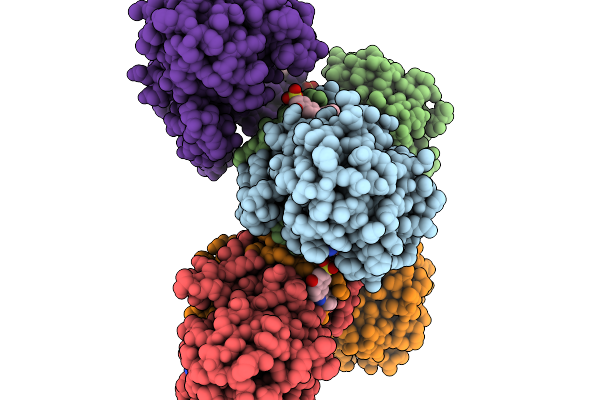 |
Structure Of Pycr1 Complexed With The Allosteric Inhibitor 4-Hydroxy-7-(Phenylamino)Naphthalene-2-Sulfonic Acid In A Remote Site And Nadh And (S)-(-)-2-Hydroxy-3,3-Dimethylbutyric Acid In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: G8I, A1CFI, NAI, PEG |
 |
Structure Of Pycr1 Complexed With The Allosteric Inhibitor 7-Benzamido-4-Hydroxynaphthalene-2-Sulfonic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: SO4, PEG, A1CFH, PGE |

