Search Count: 29,900
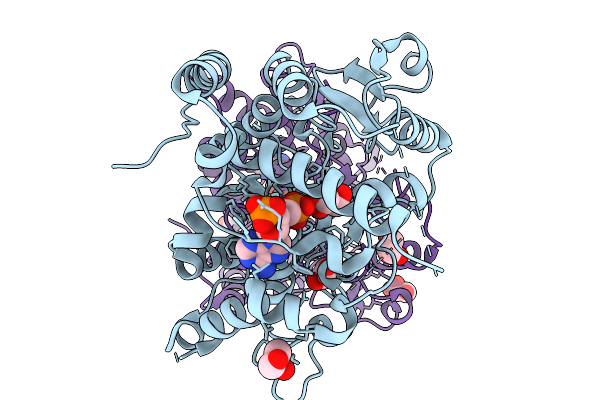 |
Crystal Structure Of Sorghum Sulfotransferase Lgs1 Reveals Sulfation-Assisted Bc-Ring Formation In Strigolactone Biosynthesis
Organism: Sorghum bicolor
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: A3P, GOL |
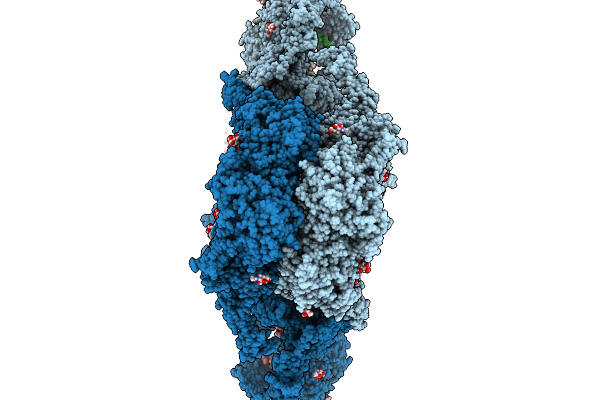 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: ENDOCYTOSIS Ligands: NAG, NGA, CA, NI |
 |
Mouse Phosphomannomutase 2 In Complex With The Activator Glucose-1,6-Bisphosphate
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2026-02-04 Classification: ISOMERASE Ligands: GOL, MG, CL, NA, G16 |
 |
Organism: Dioscoreophyllum cumminsii
Method: X-RAY DIFFRACTION Resolution:2.87 Å Release Date: 2026-02-04 Classification: PLANT PROTEIN |
 |
Structure Of Glyoxal Oxidase From Fusarium Graminearum At 1.28 Angstroms Resolution
Organism: Fusarium graminearum ph-1
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2026-02-04 Classification: OXIDOREDUCTASE Ligands: NAG, CU, SO4, NA |
 |
Three-Dimensional Structure Of Kinase Inhibitor Palbociclib-Hiv Tar Complex
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2026-02-04 Classification: RNA Ligands: LQQ |
 |
Three-Dimensional Structure Of Kinase Inhibitor Palbociclib-Hiv Tar Complex
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2026-02-04 Classification: RNA Ligands: LQQ |
 |
Three-Dimensional Structure Of Kinase Inhibitor Palbociclib-Hiv Tar Complex
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2026-02-04 Classification: RNA Ligands: LQQ |
 |
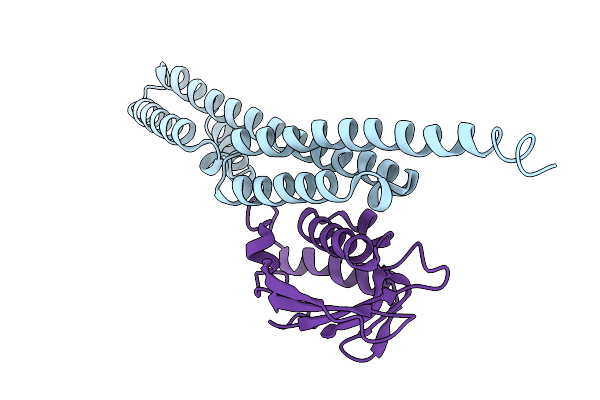
Organism: Borreliella burgdorferi b31
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN |
 |
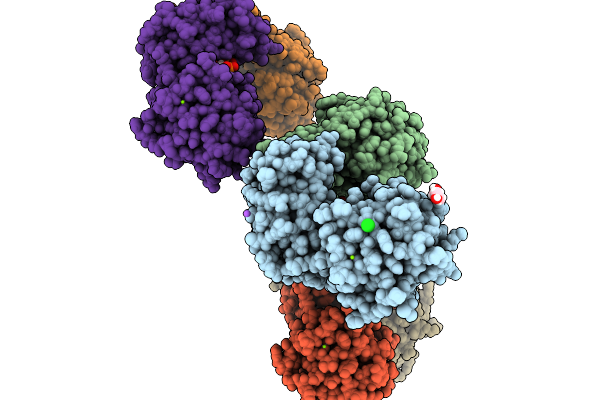
Crystal Structure Of Borrelia Burgdorferi Bb0238-Bb0323 Complex (Bb0238 Residues 118-256; Bb0323 Residues 26-210)
Organism: Borreliella burgdorferi b31
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2026-02-04 Classification: STRUCTURAL PROTEIN |
 |
Structural Characterisation Of Chromatin Remodelling Intermediates Supports Linker Dna Dependent Product Inhibition As A Mechanism For Nucleosome Spacing.
Organism: Xenopus laevis, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2026-02-04 Classification: GENE REGULATION |
 |
Human Tead1 In Complex With 2-(4-Chloro-3-{3-Methyl-5-[4-(Trifluoromethyl)Phenoxy]Phenyl}-1H-Pyrrolo[3,2-C]Pyridin-1-Yl)Ethan-1-Ol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2026-02-04 Classification: TRANSCRIPTION Ligands: MYR, GOL, CL, A1JLZ |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: NAP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: CYTOSOLIC PROTEIN Ligands: NAP |
 |
Cryo-Em Structure Of Human Udp-N-Acetylglucosamine-Dolichyl-Phosphate N-Acetylglucosaminephosphotransferase (Dpagt1) In Complex With Appb
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: A1C3G, A1C3H |
 |
Organism: Hydrogenivirga sp. 128-5-r1-1
Method: ELECTRON MICROSCOPY Resolution:2.91 Å Release Date: 2026-02-04 Classification: TRANSFERASE Ligands: A1C3G |
 |
Cryoem Structure Of Aldehyde Dehydrogenase From Francisella Tularensis Subsp. Tularensis At 3.03A Resolution
Organism: Francisella tularensis subsp. tularensis
Method: ELECTRON MICROSCOPY Resolution:3.03 Å Release Date: 2026-01-28 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Heterodimeric Crotoxin B From Crotalus Durissus Collilineatus
Organism: Crotalus durissus collilineatus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2026-01-28 Classification: LIGASE Ligands: ZN, A1CED |
 |
Organism: Rhodococcus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: OXIDOREDUCTASE Ligands: NO3, PG0 |

