Search Count: 4,024
 |
Organism: Spbetavirus spbeta
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RECOMBINATION Ligands: ZN |
 |
Organism: Spbetavirus spbeta
Method: ELECTRON MICROSCOPY Release Date: 2026-02-04 Classification: RECOMBINATION Ligands: ZN |
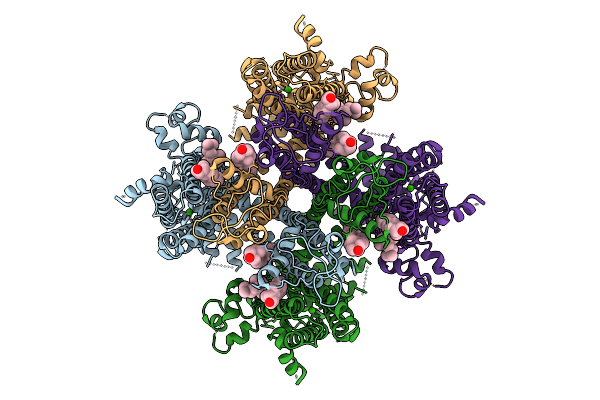 |
Human Trpm4 Ion Channel In Maasty Copolymer Lipid Nanodisc In A Calcium-Bound State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.46 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: CLR, CA |
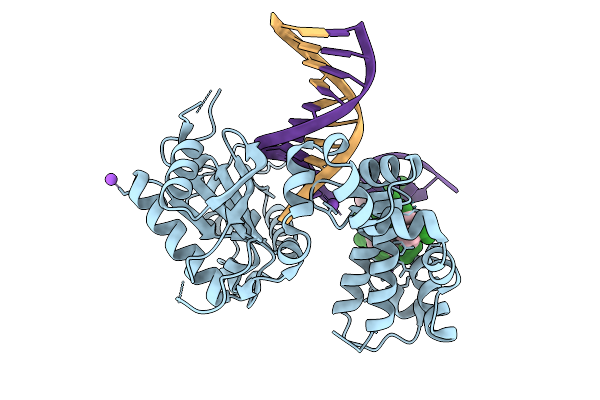 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
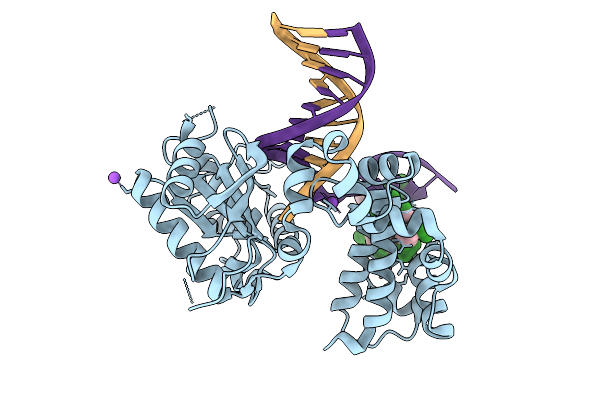 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
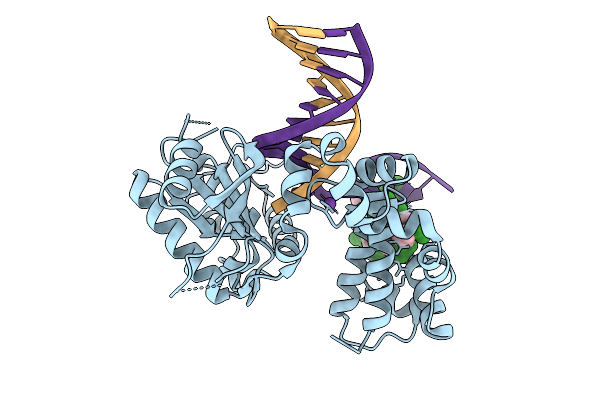 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
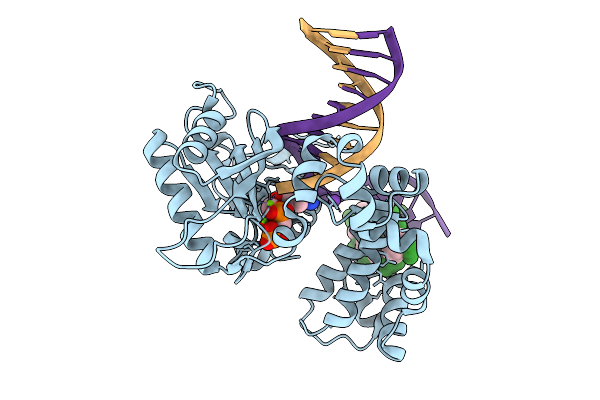 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2026-01-28 Classification: REPLICATION, HYDROLASE/DNA |
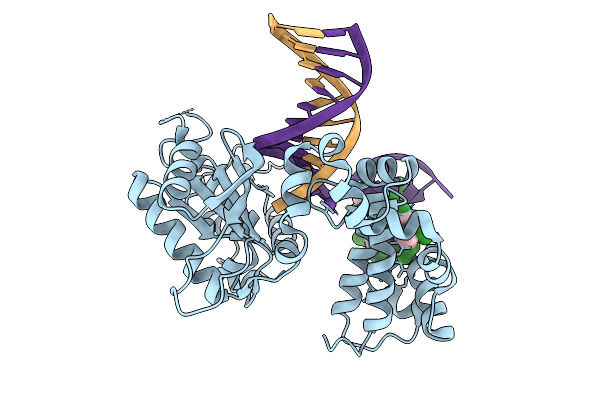 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-01-28 Classification: REPLICATION |
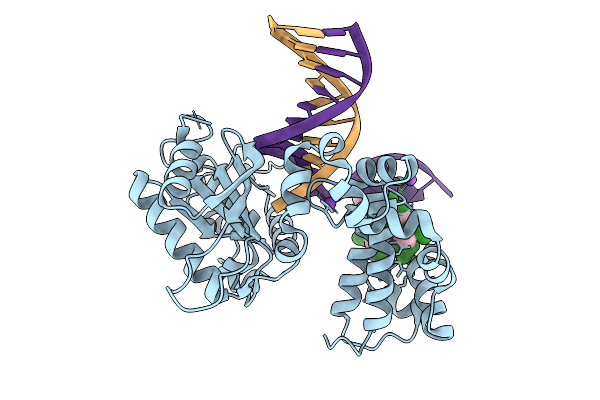 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2026-01-28 Classification: REPLICATION |
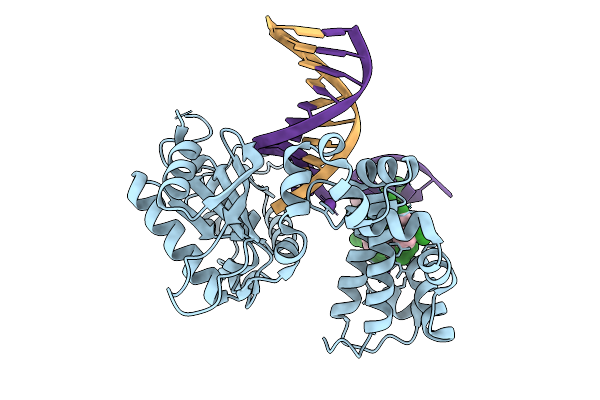 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-28 Classification: REPLICATION |
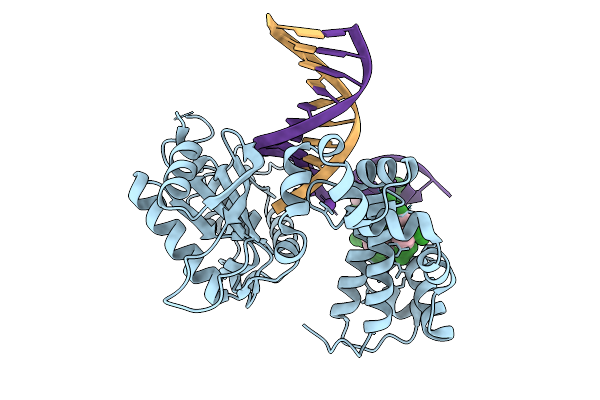 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2026-01-28 Classification: REPLICATION |
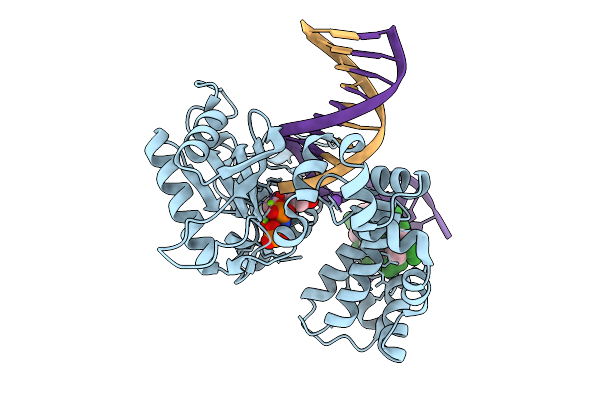 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2026-01-28 Classification: REPLICATION |
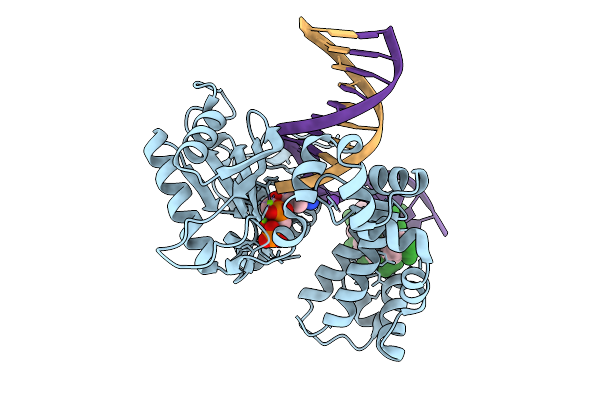 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2026-01-28 Classification: REPLICATION |
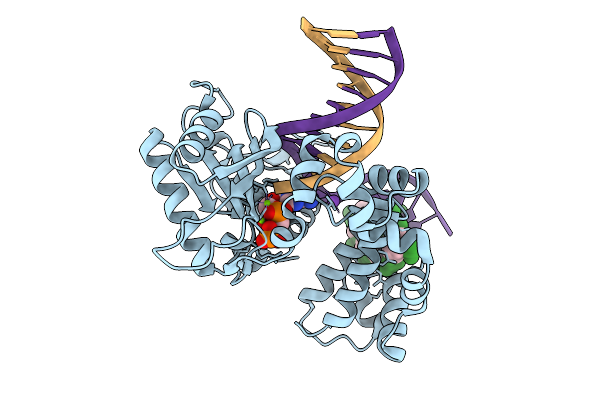 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2026-01-28 Classification: REPLICATION |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2026-01-28 Classification: REPLICATION |
 |
Organism: Escherichia coli bl21(de3), Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: TRANSLOCASE Ligands: ATP, MG |

