Search Count: 21,746
 |
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2026-01-14 Classification: DE NOVO PROTEIN Ligands: OH, DEU |
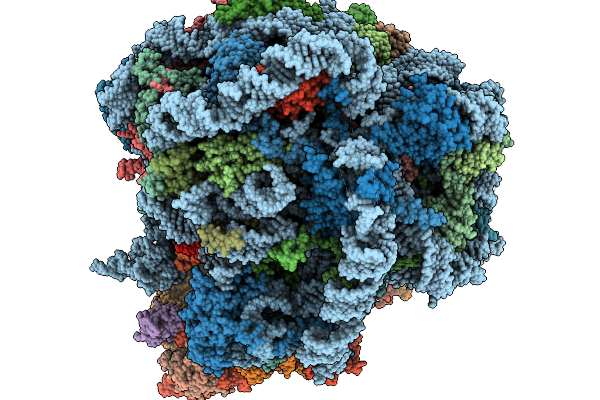 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME Ligands: MG, ZN, A1L8F |
 |
Identification And Non-Clinical Characterization Of Sar444200, A Novel Anti-Gpc3 T-Cell Engager For The Treatment Of Gpc3+ Solid Tumors
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: IMMUNE SYSTEM Ligands: NAG |
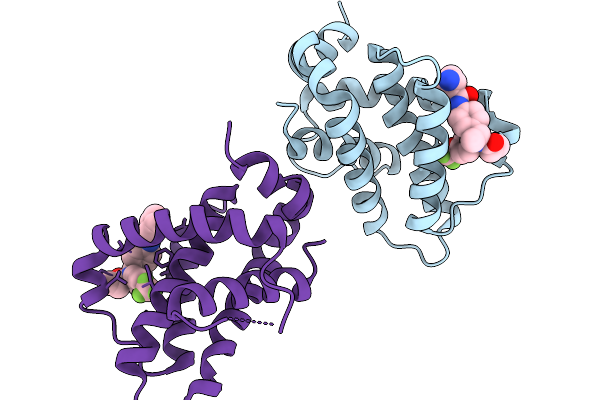 |
Structure Of Bfl1 In Complex With A Covalent Inhibitor, Alternative Series, Cmpd25
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: APOPTOSIS Ligands: A1JL2 |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: PROTEIN BINDING Ligands: CA, CL |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: PROTEIN BINDING Ligands: CL, CA |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: PROTEIN BINDING Ligands: CA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MOTOR PROTEIN Ligands: ADP, MG |
 |
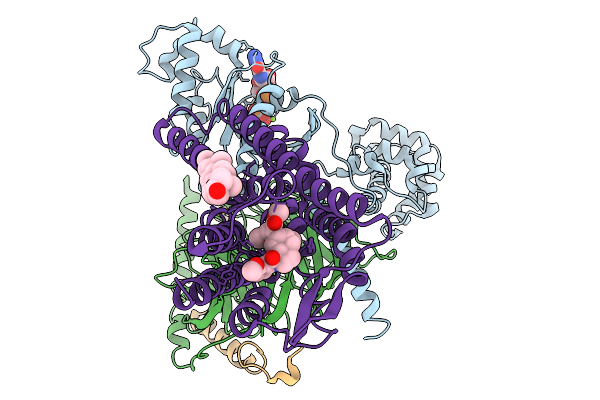
Helical Reconstruction Of The Complex Of Pseudo-Acetylated Human Cardiac Actin (K326/328Q) And Tropomyosin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: MOTOR PROTEIN Ligands: ADP, MG |
 |
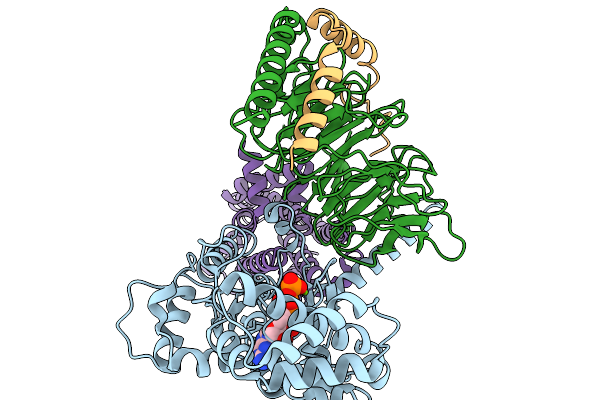
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Primed, Ahd 3Dva Sorted
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: CLR, EIG, GTP, MG |
 |
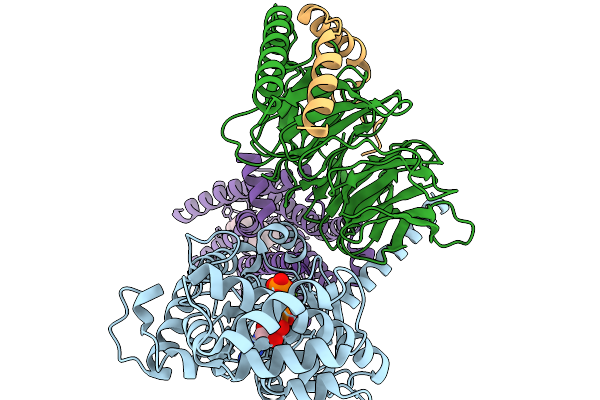
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Act-2
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: MG, GTP |
 |
Structure Of The Mor/Gi/Mitragynine Pseudoindoxil Complex, Gtp-Bound G-Act-3
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: MEMBRANE PROTEIN Ligands: MG, GTP, EIG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1C0G, SO4 |
 |
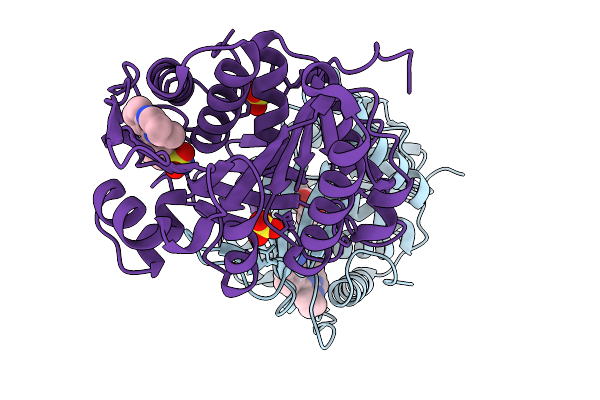
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1C0I, FRU, SO4, GOL |
 |
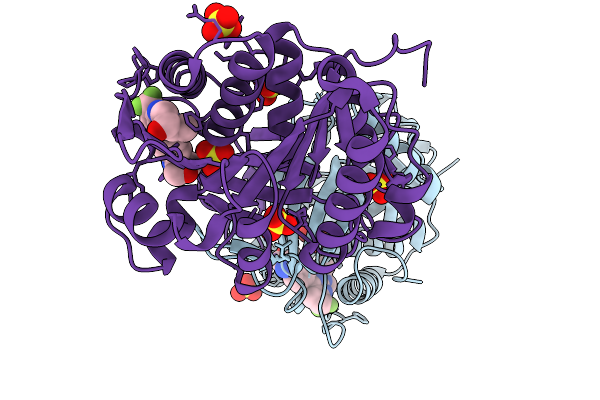
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1C0K, FRU, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1C0J, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1C0H, FRU |
 |
Cryo-Em Structure Of Violaxanthin-Chlorophyll-A-Binding Protein With Red Shifted Chl A (Rvcp) From Trachydiscus Minutus At 2.4 Angstrom
Organism: Trachydiscus minutus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, A1L1F, XAT |
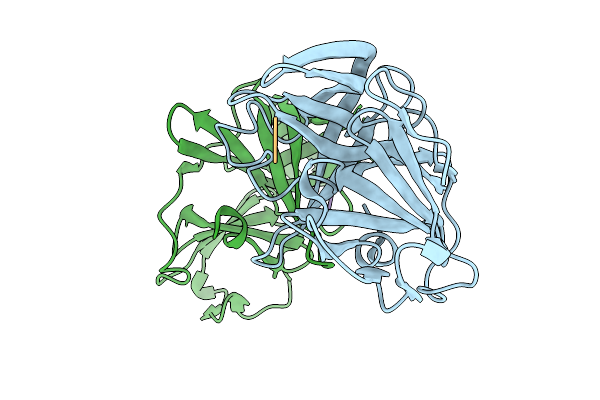 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |

