Search Count: 21,692
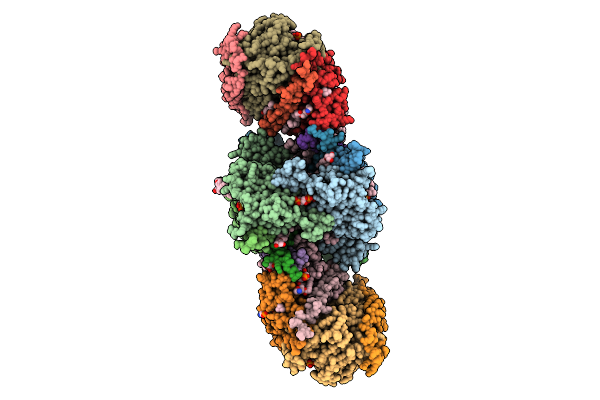 |
Organism: Paracoccus denitrificans pd1222
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: ELECTRON TRANSPORT Ligands: PC1, U10, HEM, CA, HEC, 3PE, DU0, FES, 3PH, CDL, HEA, CU, MN, CUA, ZN |
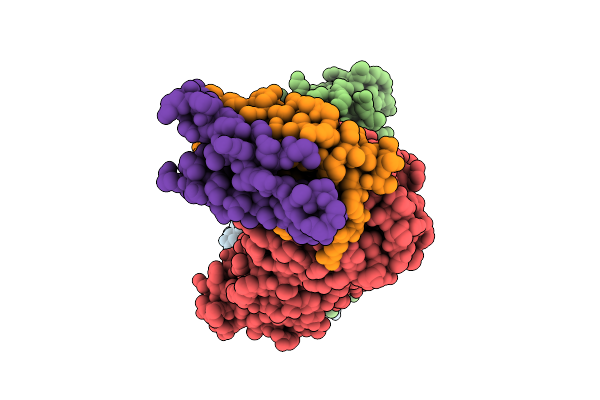 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN |
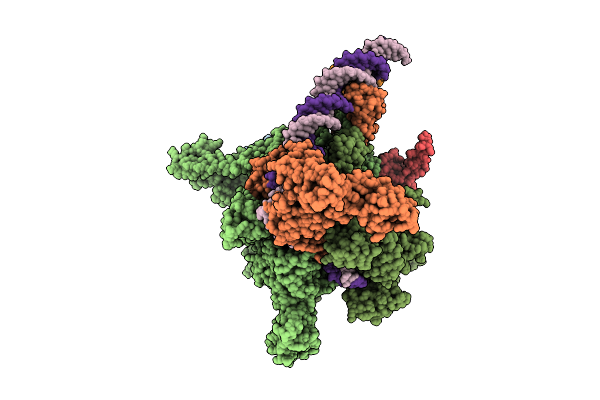 |
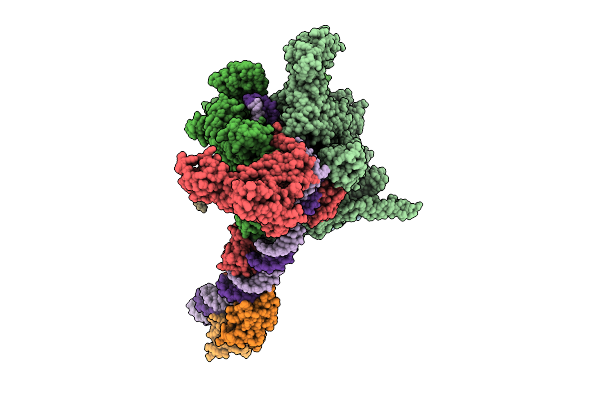
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
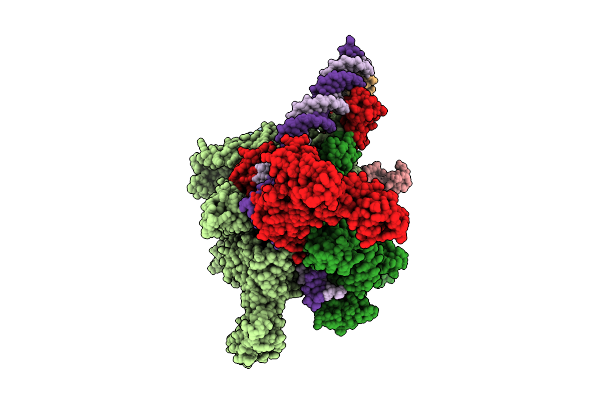 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment And 5-Mer Rna
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
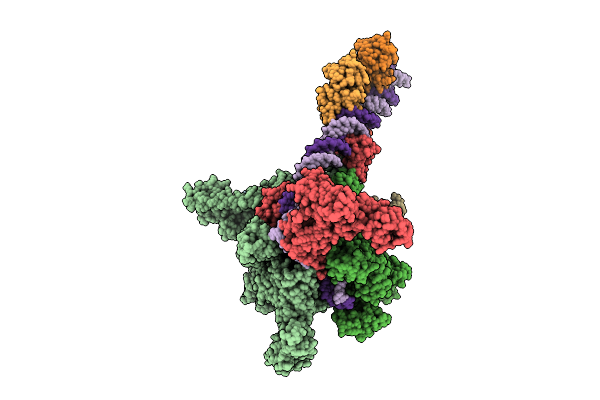
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr Transcription Factor And Ompu Promoter Dna
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
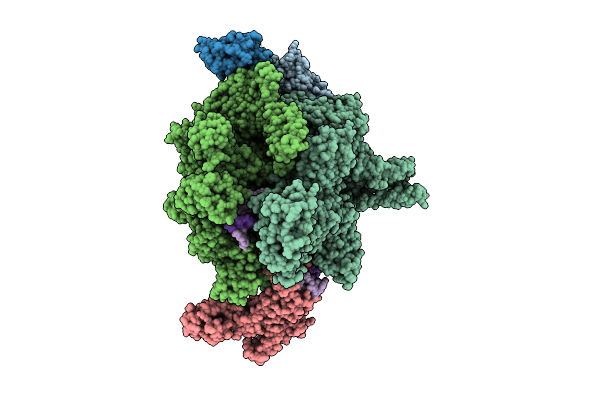
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Tcpp Transcription Factor And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
 |
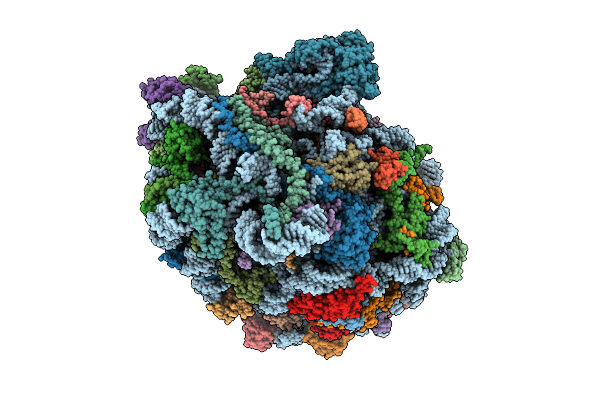
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr And Tcpp Transcription Factors And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
High Resolution Structure Of The Thermophilic 60S Ribosomal Subunit Of Chaetomium Thermophilum
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, SPM, MG, K, ZN |
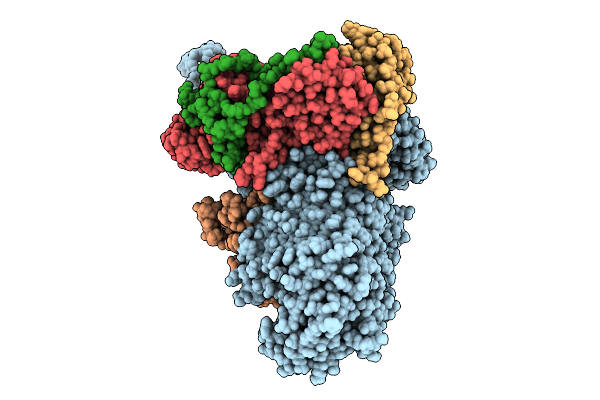 |
Beta-Barrel Assembly Machine From Escherichia Coli In An Early State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
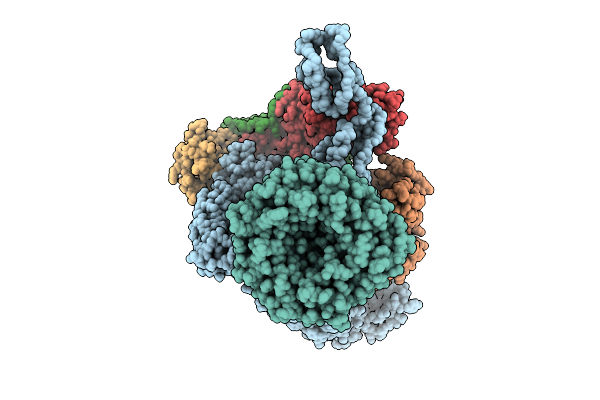 |
Beta-Barrel Assembly Machine From Escherichia Coli In A Middle State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
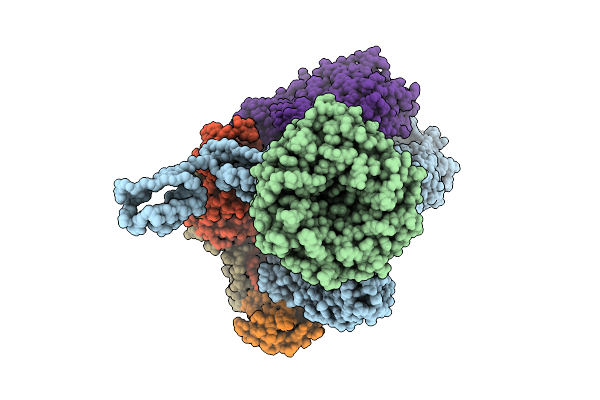 |
Beta-Barrel Assembly Machine From Escherichia Coli In A Late State Of Substrate Assembly
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Vibrio cholerae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DNA BINDING PROTEIN/DNA |
 |
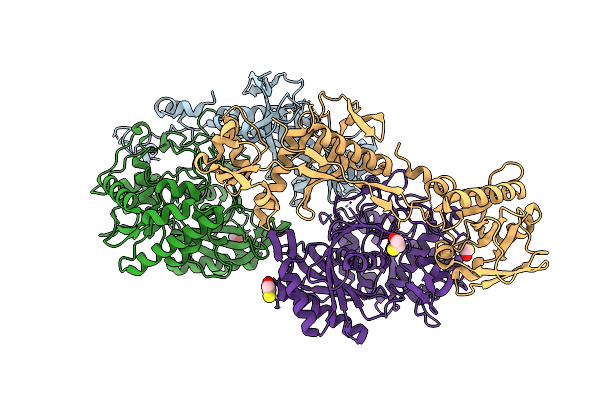
Crystal Structure Of Xanthobacter Autotrophicus Sparda Mutant Lacking Dren Nuclease Domains
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: ACY, GOL, TRS, PEG, CL, BME, TAR |
 |
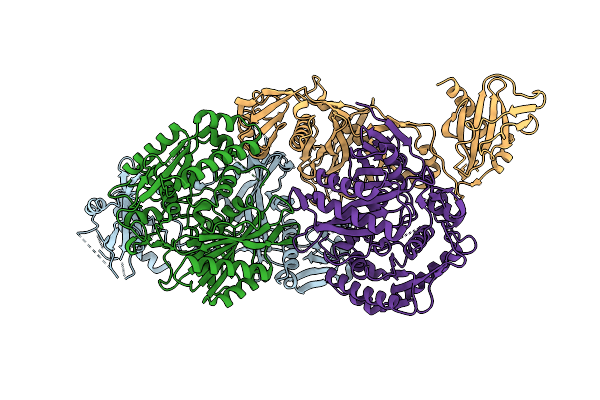
Organism: Enhydrobacter aerosaccus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: CL, TRS, BME |
 |
Organism: Xanthobacter autotrophicus py2
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: SO4 |
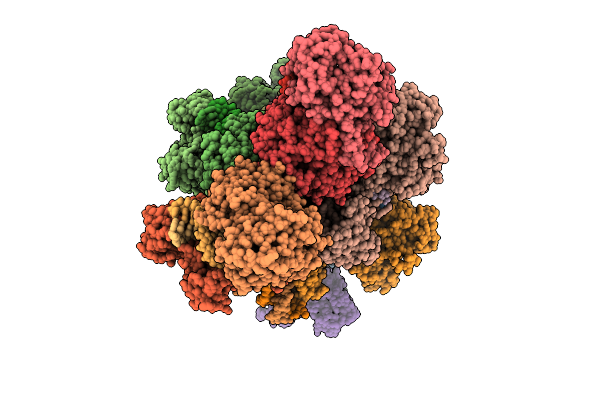 |
Organism: Xanthobacter autotrophicus py2, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RNA BINDING PROTEIN Ligands: CA |
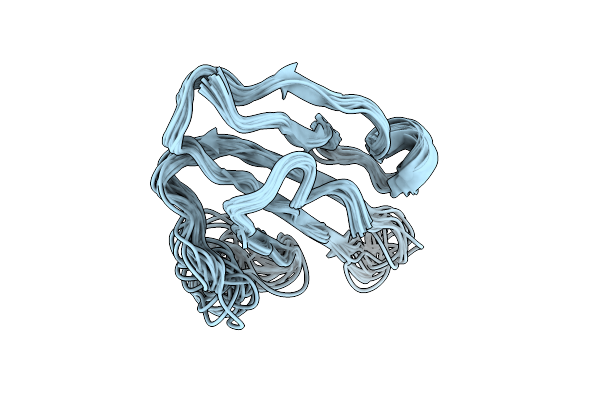 |
Solution Structure Of The Human Prostate Stem Cell Antigen (Psca), Water-Soluble Domain
|

