Search Count: 14,894
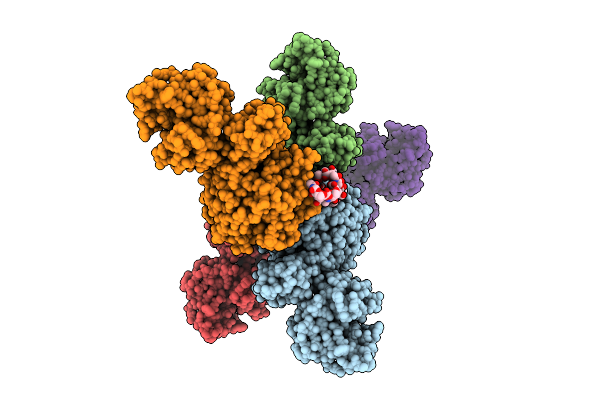 |
Organism: Streptomyces coelicolor
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TOXIN Ligands: A2G, A1CAY, A1CAZ |
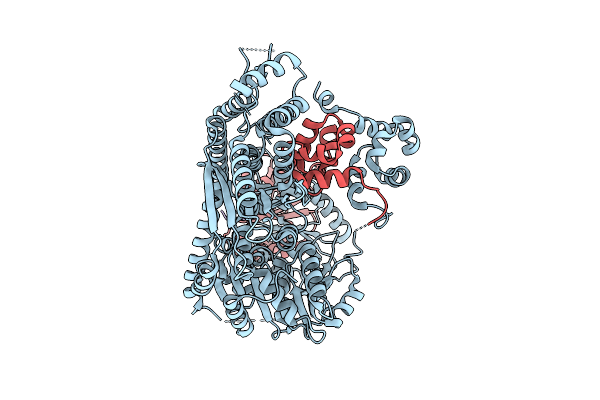 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE |
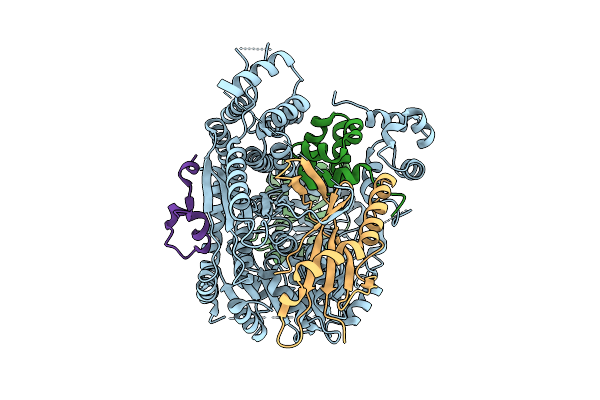 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE |
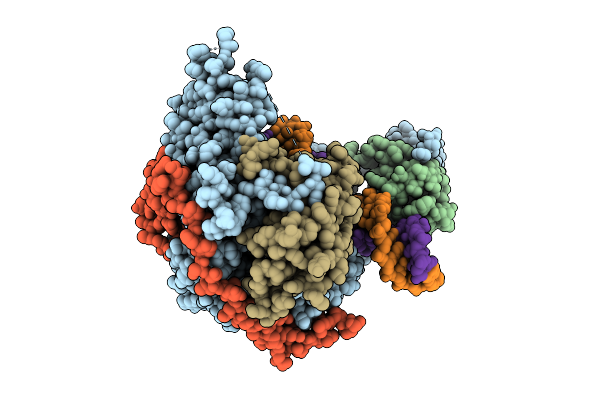 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: HYDROLASE Ligands: MN |
 |
Cryo-Em Structure Of Spinacia Oleracea Cytochrome B6F Complex With Bound Plastocyanin
Organism: Spinacia oleracea
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: OXIDOREDUCTASE Ligands: HEM, HEC, UMQ, PL9, CLA, FES, SQD, BCR, CU |
 |
Organism: Homo sapiens, Lama glama, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: DE NOVO PROTEIN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: LIGASE |
 |
M. Tuberculosis Clpc1-Ntd Complexed With A Click Chemistry Analog Of Rufomycin
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIBIOTIC |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GDP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GTP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE Ligands: MG, GNP |
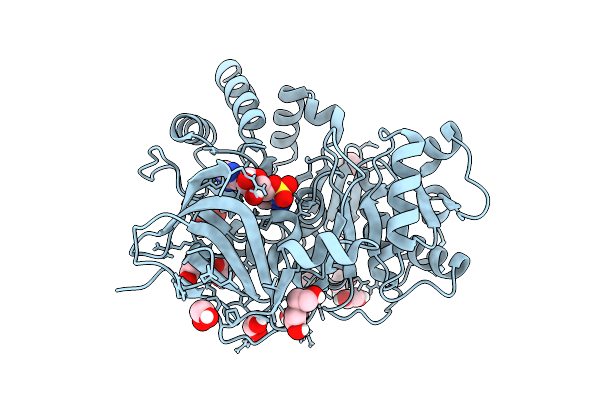 |
High-Resolution Crystal Structure Of Methyl-2,3-Diamino Propanoic Acid-Ams Inhibitor Bound Adenylation Domain (A3) From Sulfazecin Nonribosomal Peptide Synthetase Sulm
Organism: Paraburkholderia acidicola
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: LIGASE Ligands: A1BVA, EDO, PEG |
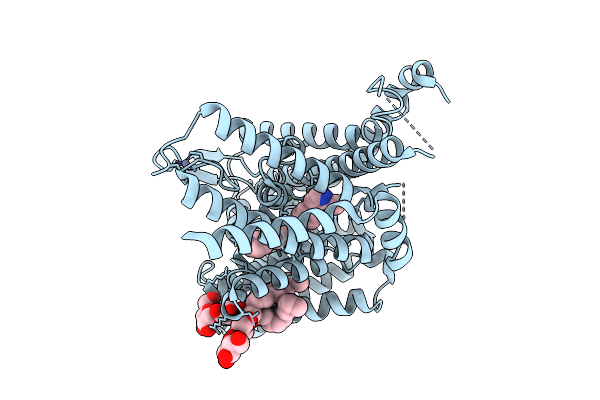 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CC5, AV0, ZN |
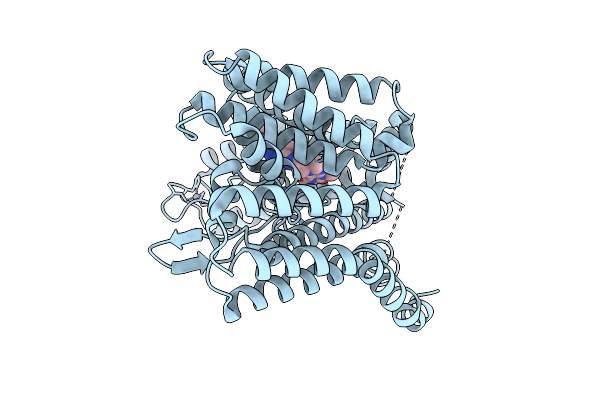 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ZN, A1CC6 |
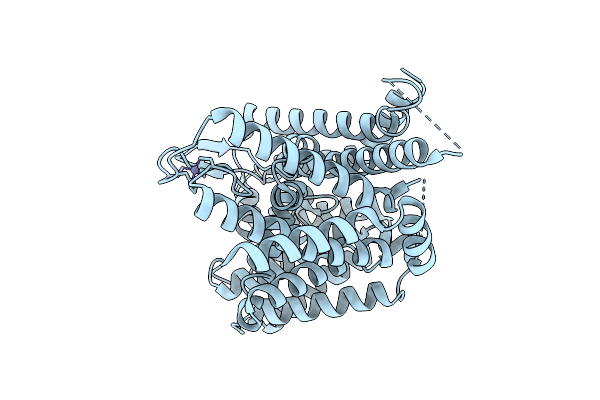 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: ZN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN Ligands: PGE, NA |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN Ligands: GOL, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: OXIDOREDUCTASE Ligands: A1JLE, SO4, PG4, PGE, PEG |

