Search Count: 30,199
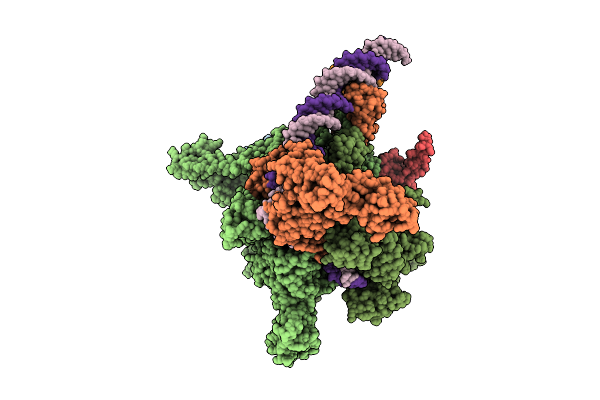 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
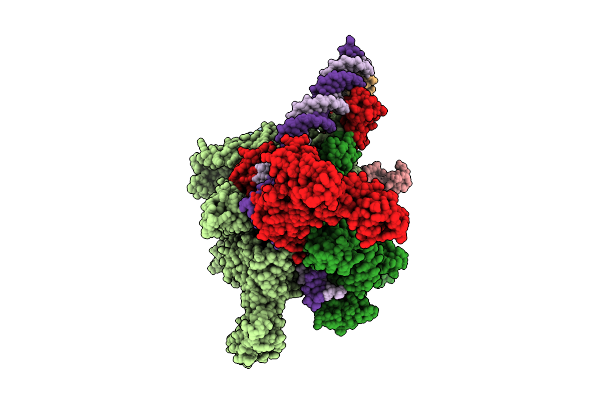 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Holoenzyme Bound To An Ompu Promoter Dna Fragment And 5-Mer Rna
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
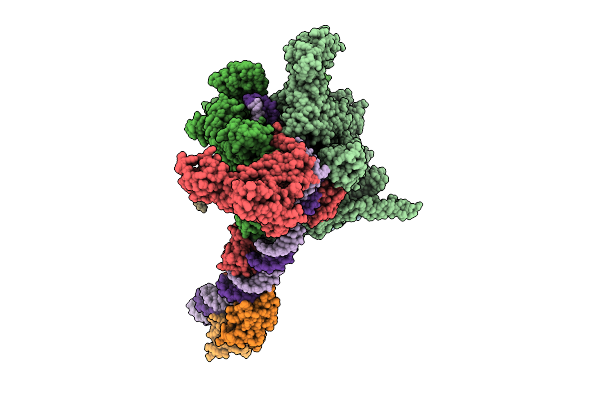 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr Transcription Factor And Ompu Promoter Dna
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
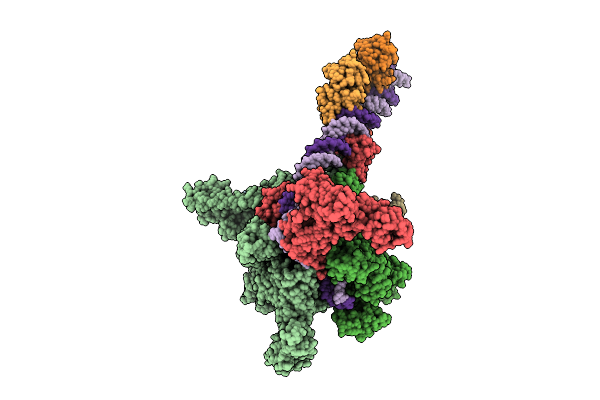 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Tcpp Transcription Factor And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: ZN, MG |
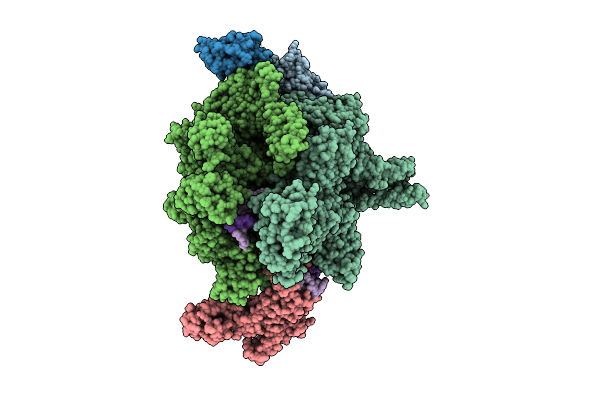 |
Cryo-Em Structure Of Vibrio Cholerae Rna Polymerase Transcription Activation Complex With Toxr And Tcpp Transcription Factors And A Toxt Promoter Dna Fragment
Organism: Vibrio cholerae o395, Vibrio cholerae o1 biovar el tor str. n16961
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: MG, ZN |
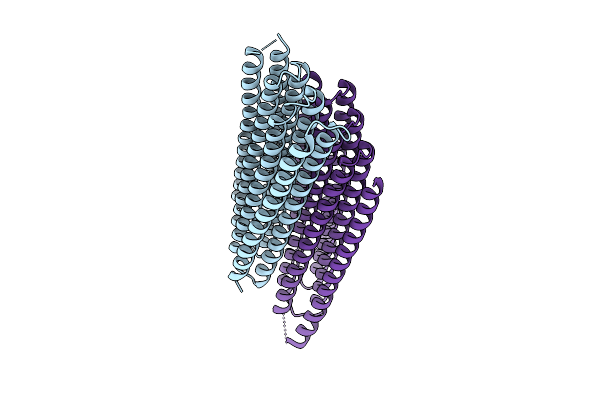 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: DE NOVO PROTEIN |
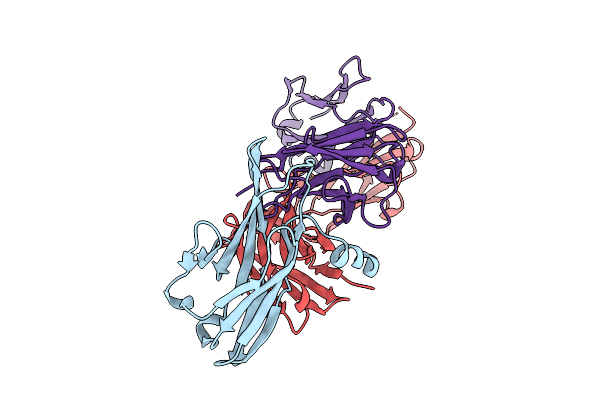 |
Crystal Structure Of Pilra In Complex With Fab Portion Of Antagonist Antibody
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: SIGNALING PROTEIN |
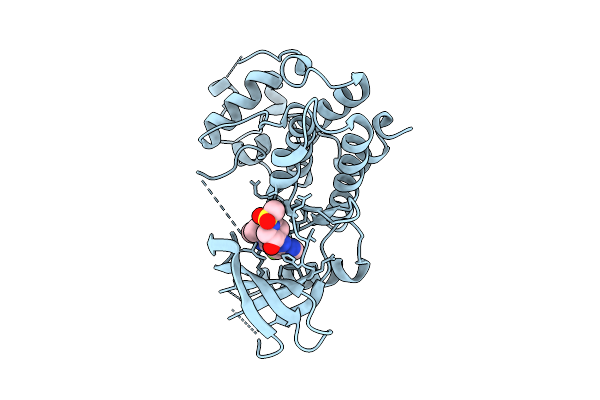 |
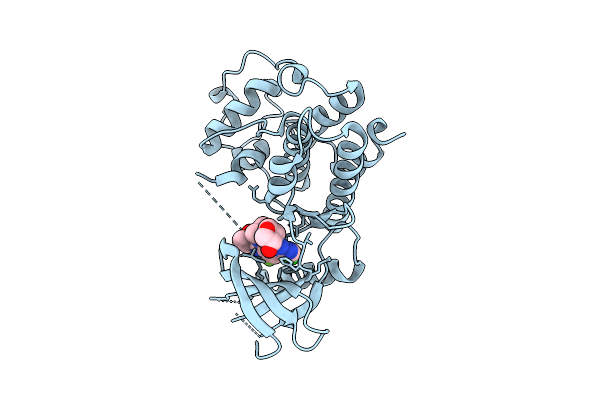
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1CIF |
 |
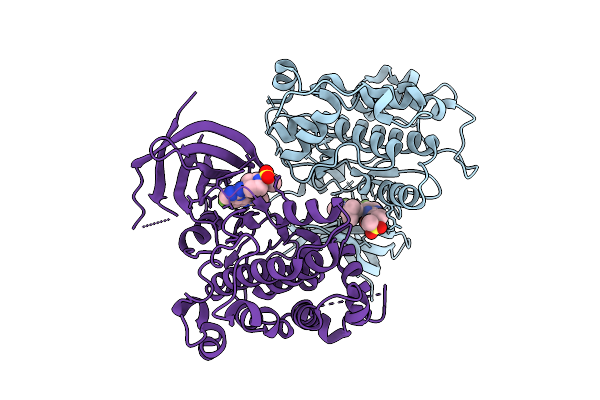
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1AZ4 |
 |
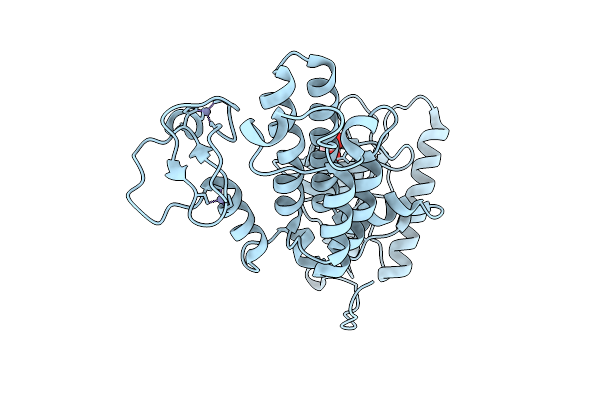
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSFERASE/INHIBITOR Ligands: A1CIF |
 |
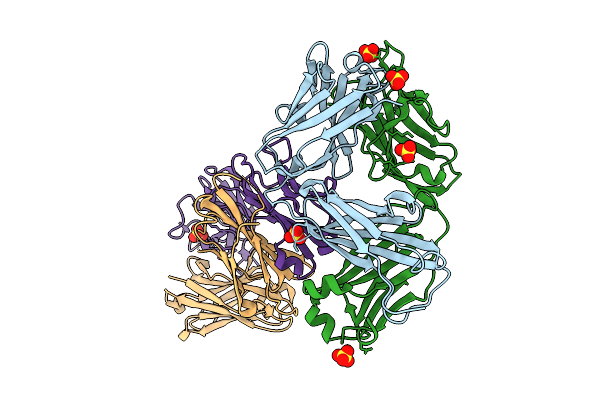
Crystal Structure Of Human Prkcbp1 Zinc Finger Mynd-Type Containing 8 With Pentaglutamate Tag
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: TRANSCRIPTION Ligands: EDO, ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM Ligands: SO4 |
 |
Organism: Mus musculus, Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
M. Tuberculosis Clpc1-Ntd Complexed With A Click Chemistry Analog Of Rufomycin
Organism: Mycobacterium tuberculosis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: ANTIBIOTIC |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: DE NOVO PROTEIN |
 |
A Rare Open Conformation For Ubl2 Domain Of Papain-Like Protease C111S Of Sars-Cov2
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: VIRAL PROTEIN Ligands: GOL, ZN, SO4 |
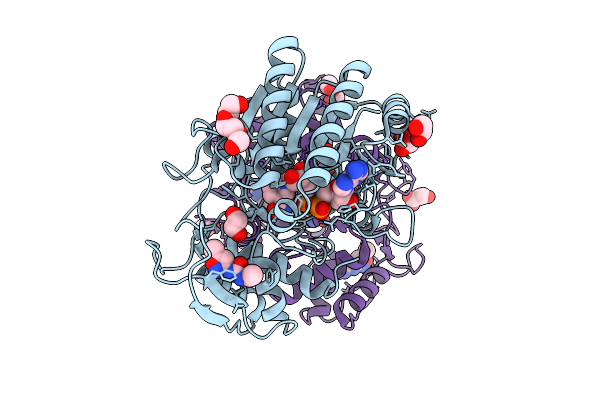 |
Structure Of Udp-Galactose-4-Epimerase (Gale) Bound To Fragment From Diamond Xchem Experiment.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CARBOHYDRATE Ligands: NAD, JGA, MLI, PGE, EDO, CL |
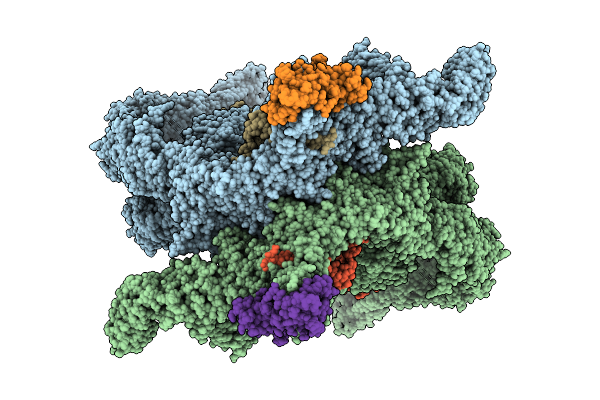 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN |
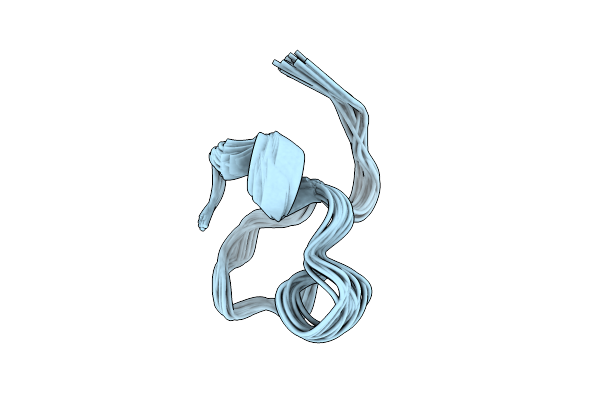 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
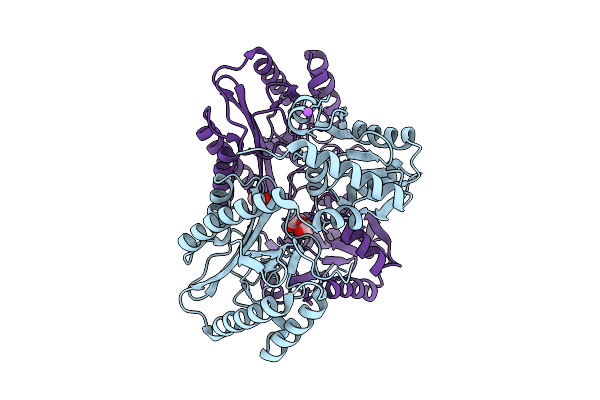 |
Crystal Structure Of Proline Dipeptidase From Pyrococcus Furiosus Complexed With Co
Organism: Pyrococcus furiosus dsm 3638
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE |

