Search Count: 3,728
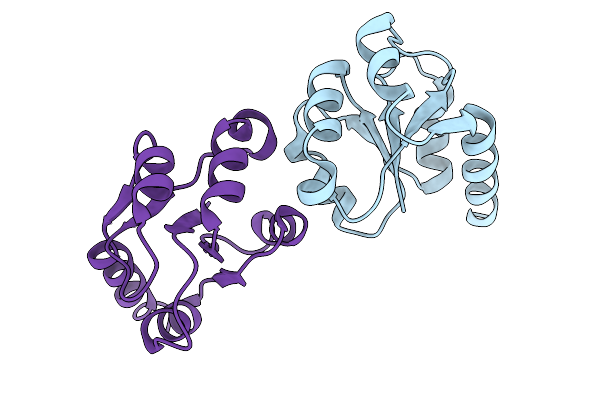 |
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
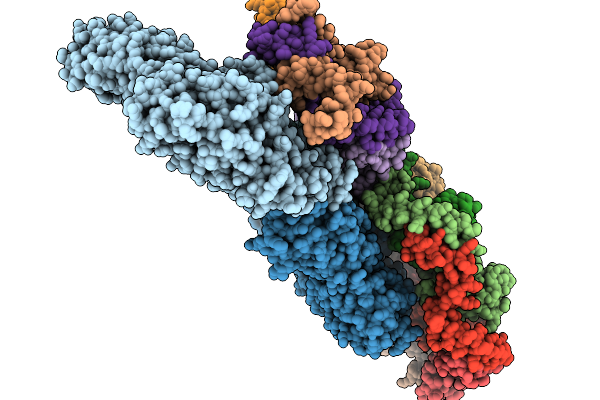 |
Asymmetric Unit From Transcribing Double-Layered Particle Of Bacteriophage Phi6
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
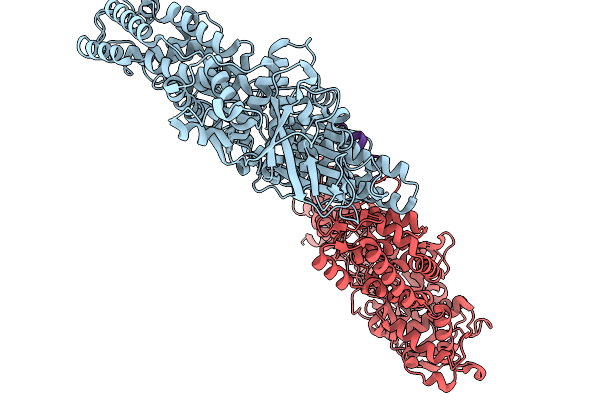 |
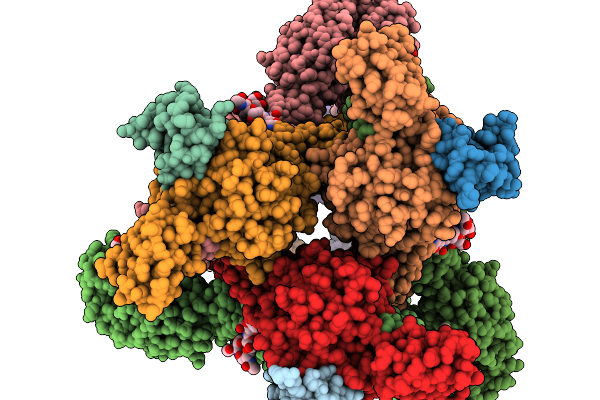
Asymmetric Unit From Transcribing Single-Layered Particle Of Bacteriophage Phi6
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
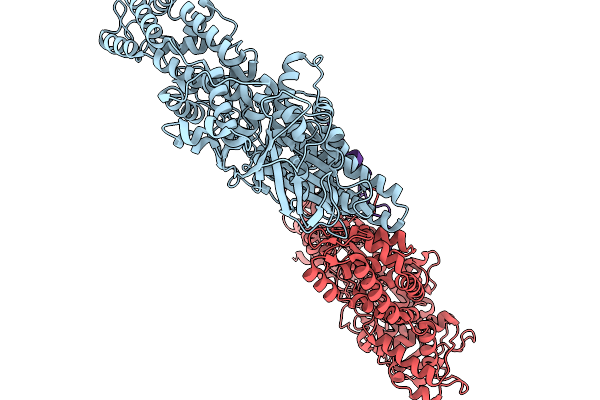 |
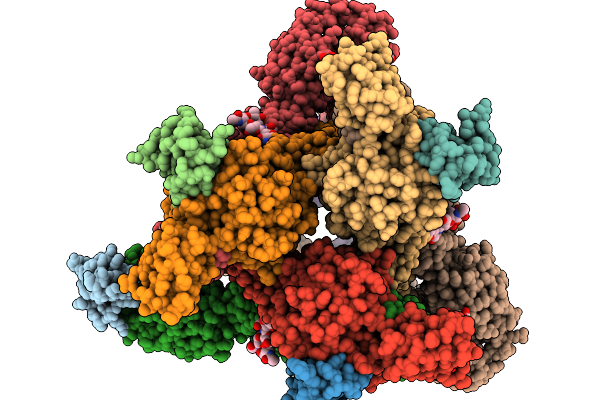
Asymmetric Unit From Transcribing Single-Layered Particle Of Bacteriophage Phi6 In An Over-Expanded State
Organism: Cystovirus phi6
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
 |
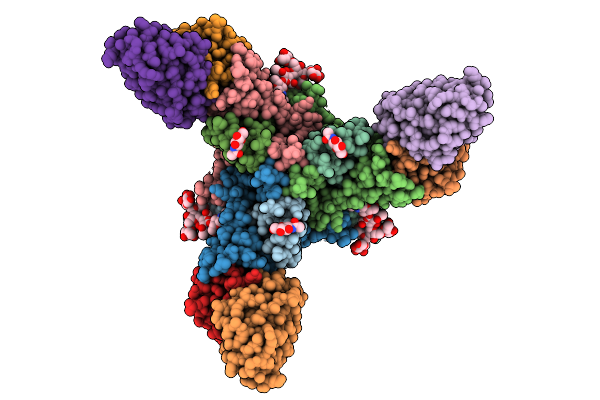
Organism: Homo sapiens, Mus musculus, Semliki forest virus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS Ligands: CA |
 |
Organism: Semliki forest virus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: VIRUS |
 |
Organism: Homo sapiens, Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: ACT, NA |
 |
Organism: Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: IPA |
 |
Organism: Homo sapiens, Bacteroides uniformis atcc 8492
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-12-03 Classification: BIOSYNTHETIC PROTEIN Ligands: CIT, PEG |
 |
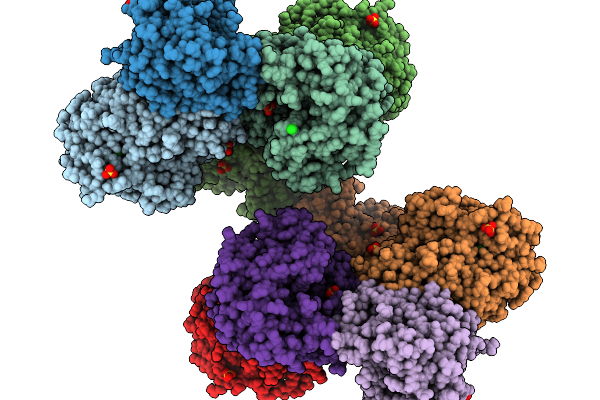
Organism: Mus musculus, Orthomarburgvirus marburgense
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2025-11-26 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Thermomicrobium roseum dsm 5159
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: SO4, CL |
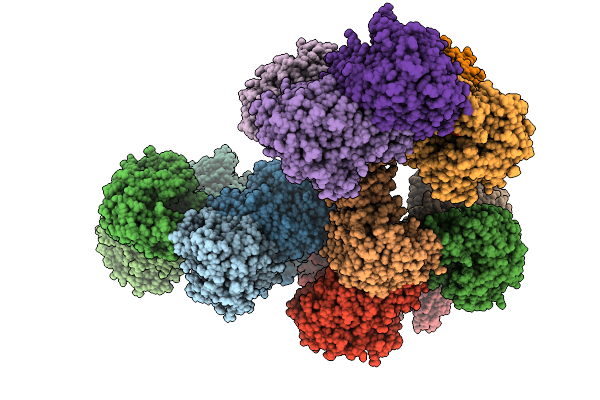 |
Organism: Thermomicrobium roseum dsm 5159
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2025-11-19 Classification: OXIDOREDUCTASE Ligands: BG6, PEG, PG4, PGE, EDO |
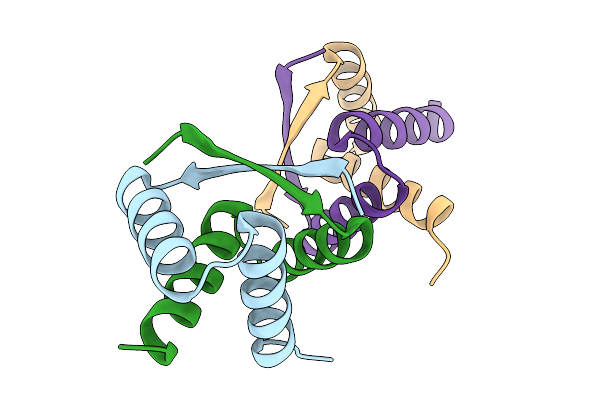 |
Organism: Streptomyces albus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
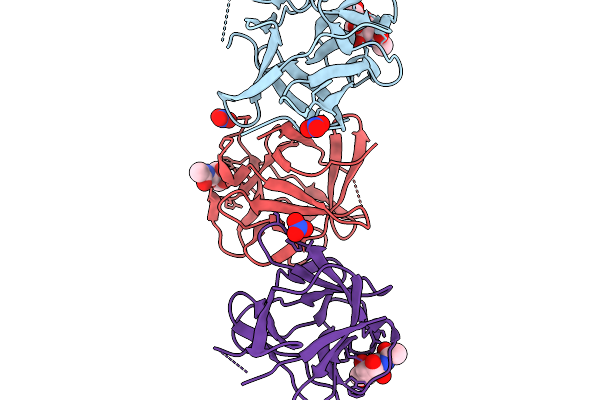 |
Crystal Structure Of Pleurocybella Porrigens Lectin (Ppl) In Complex With Galnac
Organism: Pleurocybella porrigens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-11-19 Classification: SUGAR BINDING PROTEIN Ligands: A2G, NO3 |
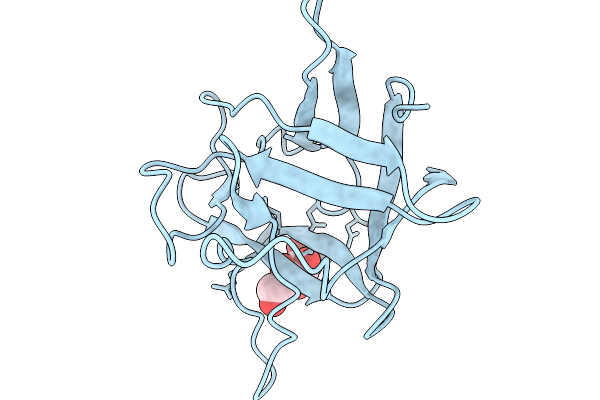 |
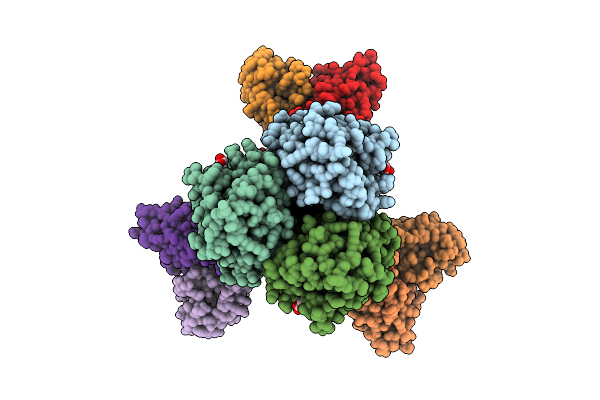
Cryoem Structure Of Pleurocybella Porrigens Lectin (Ppl) In Complex With Galnac
Organism: Pleurocybella porrigens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: SUGAR BINDING PROTEIN Ligands: A2G |
 |
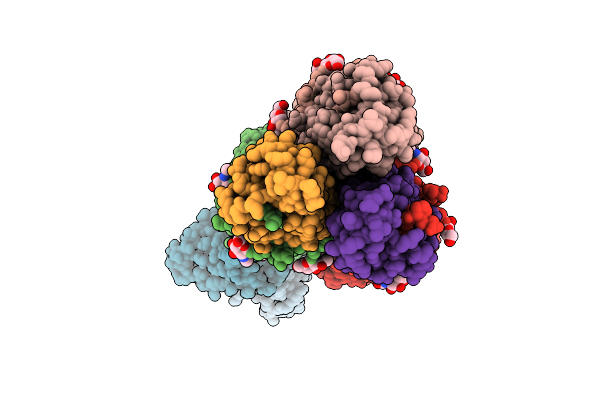
Organism: Homo sapiens, Influenza a virus (a/california/01/2009(h1n1))
Method: ELECTRON MICROSCOPY Resolution:4.10 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens, Influenza a virus (a/california/01/2009(h1n1))
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-11-12 Classification: IMMUNE SYSTEM/VIRAL PROTEIN Ligands: NAG |
 |
Cryo-Em Structure Of Broad Betacoronavirus Binding Antibody 1871 In Complex With Oc43 S2 Subunit
Organism: Human coronavirus oc43, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
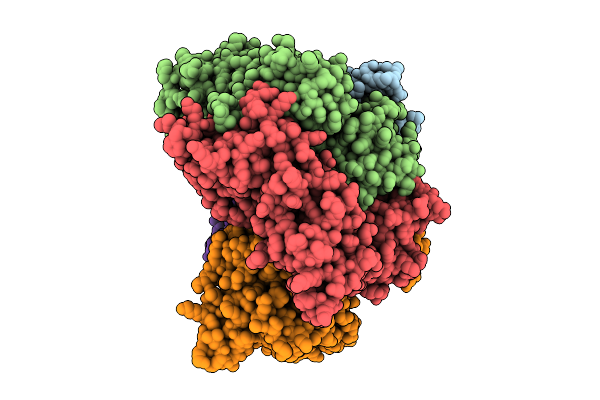 |
Structure Of The S. Cerevisiae Clamp Loader Replication Factor C (Rfc) With Mixed Nucleotide Occupancy
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: REPLICATION Ligands: ACT, ADP, GDP |
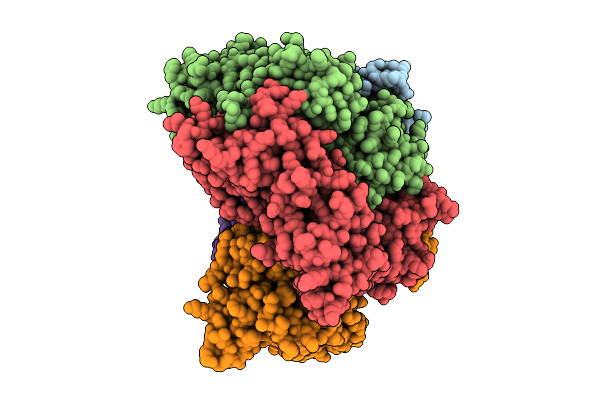 |
Structure Of The S. Cerevisiae Clamp Loader Replication Factor C (Rfc) With Mixed Nucleotide Occupancy
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: REPLICATION Ligands: ACT, ADP, GDP |

