Search Count: 15,401
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, CL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, CL, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, CL, MG |
 |
Iron Loaded Human Mitochondrial Ferritin D131N Mutant 20 Minute Oxygen Soak
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2026-01-28 Classification: METAL BINDING PROTEIN Ligands: FE, MG, CL |
 |
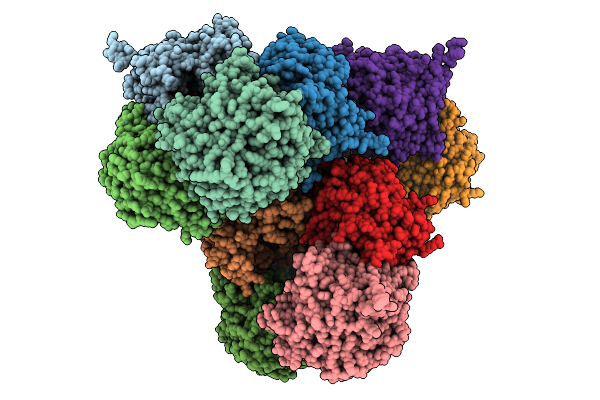
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:2.99 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: D21 |
 |
Organism: Prevotella
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2026-01-28 Classification: HYDROLASE Ligands: GOL |
 |
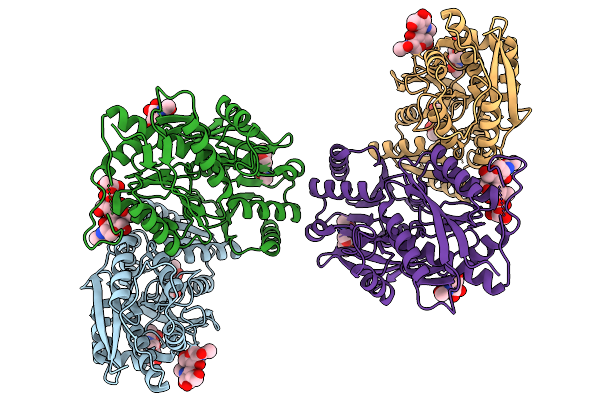
Crystal Structure Of Human Cd22 Ig Domains 1-3 In Complex With Modified Sialoside 17
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2026-01-28 Classification: IMMUNE SYSTEM Ligands: SO4, A1JJP |
 |
Organism: Sus scrofa
Method: ELECTRON MICROSCOPY Resolution:3.26 Å Release Date: 2026-01-28 Classification: SIGNALING PROTEIN Ligands: NAG |
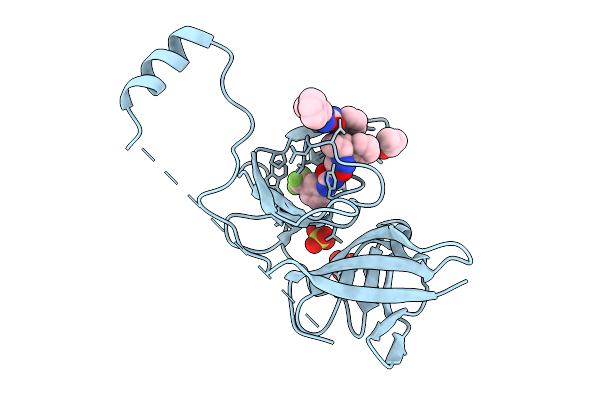 |
Crystal Structure Of Zika Virus Ns2B-Ns3 Protease In Complex With Compound 1
Organism: Zika virus
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2026-01-28 Classification: VIRAL PROTEIN Ligands: A1H2Z, SO4, IPA |
 |
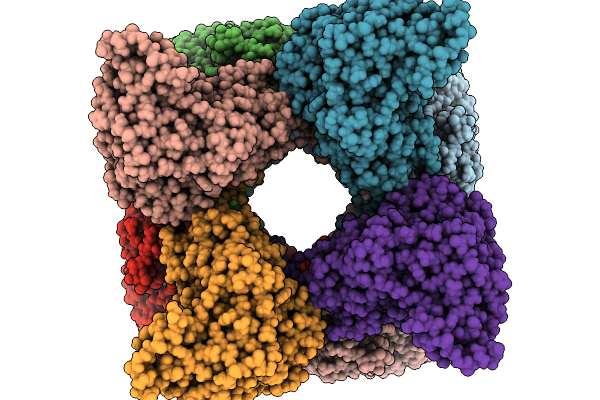
Mitochondrial Creatine Kinase In Complex With Adp, Creatine, And Uncompetitive Inhibitor Uci
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.60 Å Release Date: 2026-01-28 Classification: TRANSFERASE/INHIBITOR Ligands: ADP, CRN, A1CZQ, MG |
 |
Mitochondrial Creatine Kinase In Complex With Adp And Uncompetitive Inhibitor Uci
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.59 Å Release Date: 2026-01-28 Classification: TRANSFERASE/INHIBITOR Ligands: ADP, A1CZQ, MG |
 |
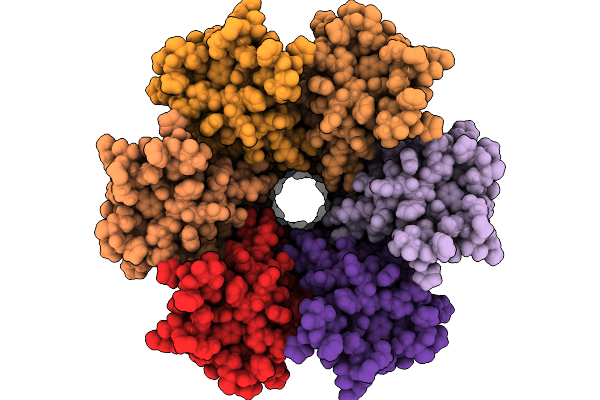
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN |
 |
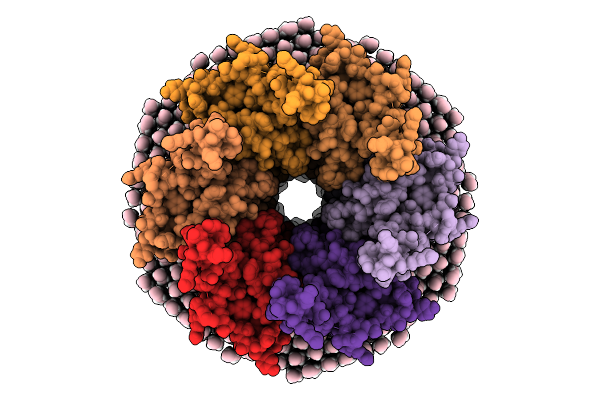
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN |
 |
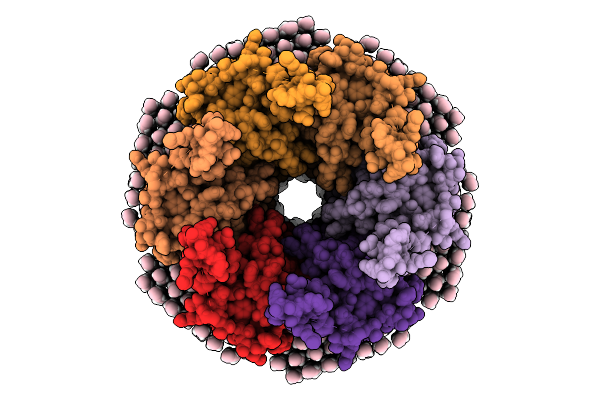
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
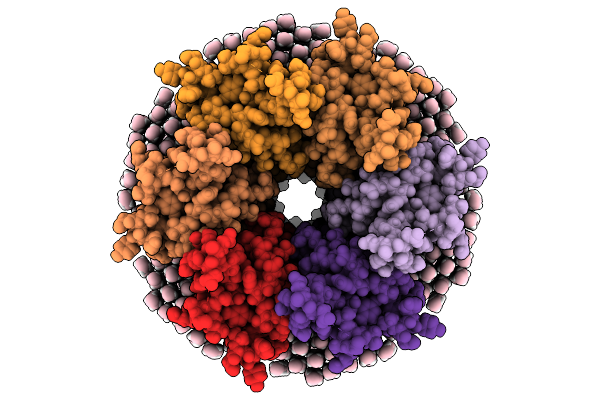
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: MC3 |
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: MC3 |

