Search Count: 5,898
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-17 Classification: PROTEIN BINDING |
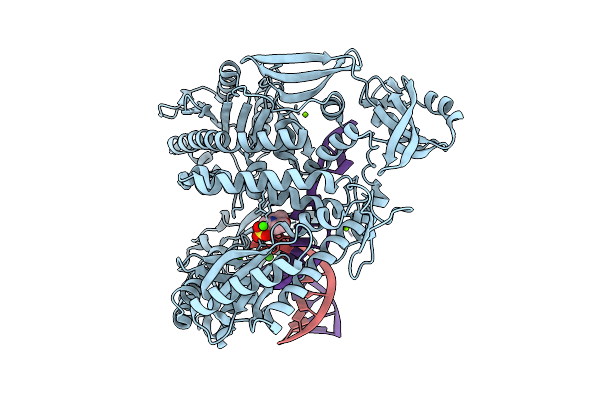 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: CA, MG, MES |
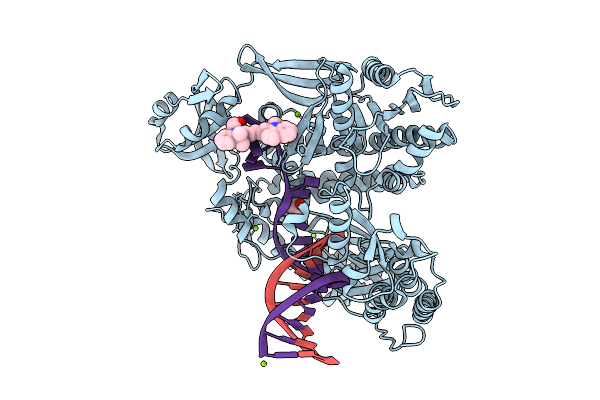 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG, 5CY, GOL |
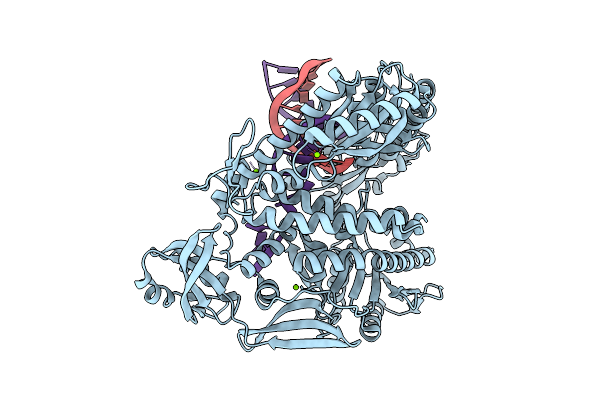 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
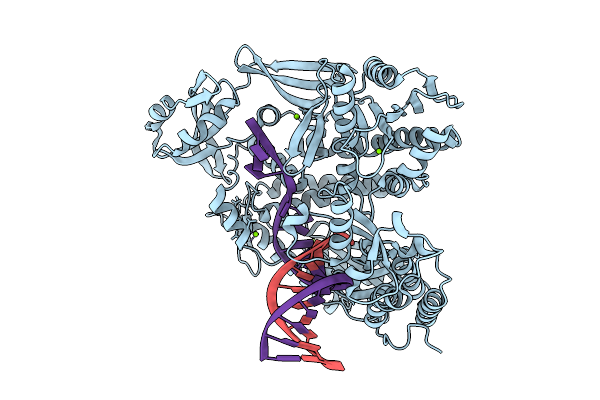 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, EDO |
 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, CL |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS Ligands: NAG |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: VIRUS |
 |
Human G Protein-Coupled Receptor Kinase 5-D311N In Complex With Sangivamycin Soaked In Ph 6
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: SIGNALING PROTEIN Ligands: SGV, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSCRIPTION/INHIBITOR Ligands: BME, 9N6, NA, GOL, SO4 |
 |
Organism: Porphyromonas gingivalis atcc 33277
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: PLM, Z41 |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN |
 |
The Crystal Structure Of Cyp125Mrca, An Ancestrally Reconstructed Cyp125 Enzyme
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: LIPID BINDING PROTEIN Ligands: HEM, VD3, EDO, IPA, NA |
 |
Crystal Structure Of Human G Protein-Coupled Receptor Kinase 5 In Complex With Grl098-22
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: A1ARA |
 |
Crystal Structure Of Human G Protein-Coupled Receptor Kinase 5 In Complex With Grl080-22
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: A1ARB |
 |
Crystal Structure Of Human G Protein-Coupled Receptor Kinase 5 In Complex With Grl077-22
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: A1ARC |
 |
Crystal Structure Of Human G Protein-Coupled Receptor Kinase 5 In Complex With Grl050-22
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: SIGNALING PROTEIN/INHIBITOR Ligands: A1ARD |

