Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2026-01-14 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2026-01-14 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: IMMUNE SYSTEM Ligands: Q87 |
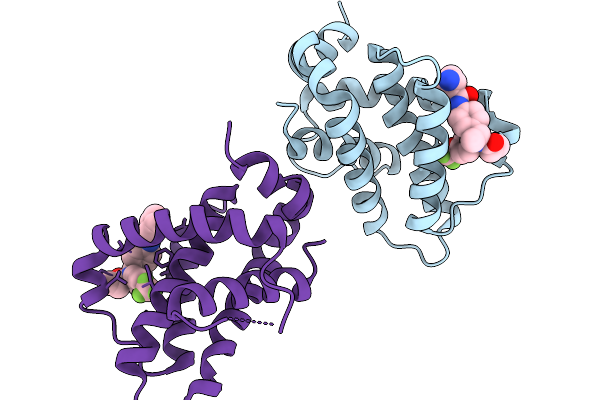 |
Structure Of Bfl1 In Complex With A Covalent Inhibitor, Alternative Series, Cmpd25
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: APOPTOSIS Ligands: A1JL2 |
 |
Human Ectonucleotide Pyrophosphatase/Phosphodiesterase Family Member 3 (Enpp3) Inhibitor Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: HYDROLASE Ligands: A1BYU, ZN, CA, NAG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1CMW, MTA |
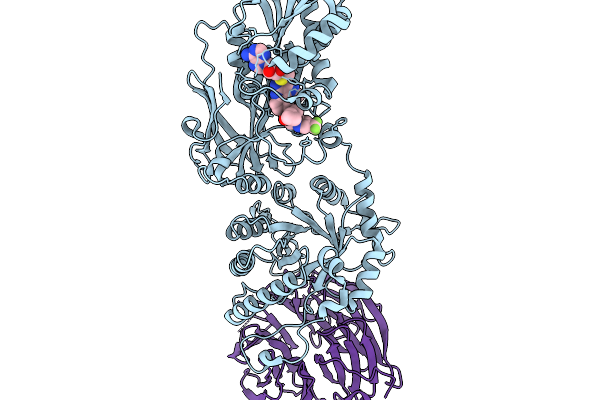 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1CMZ, MTA |
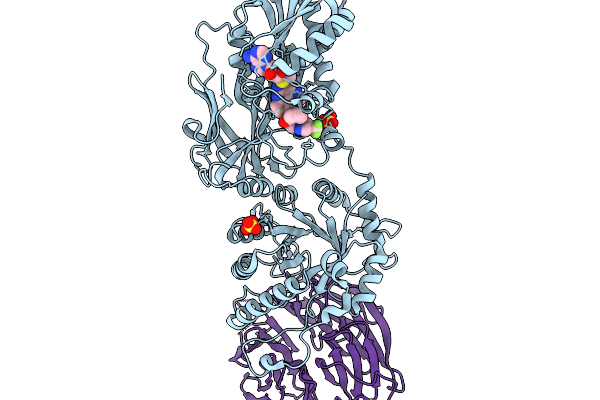 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-07 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MTA, A1CM0, SO4 |
 |
Crystal Structure Of Gitr In Complex With Ligand-Non-Competitive Ab#1 Fab Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: IMMUNE SYSTEM Ligands: ACT, NAG |
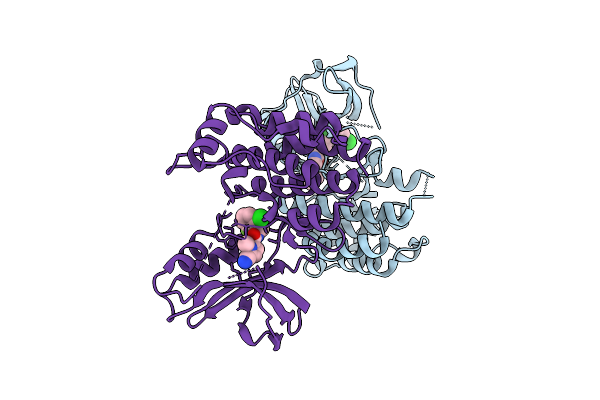 |
Crystal Structure Of An Allosteric Inhibitor Bound To Human Ripk1 Kinase Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: A1IX7, IOD, 1HX |
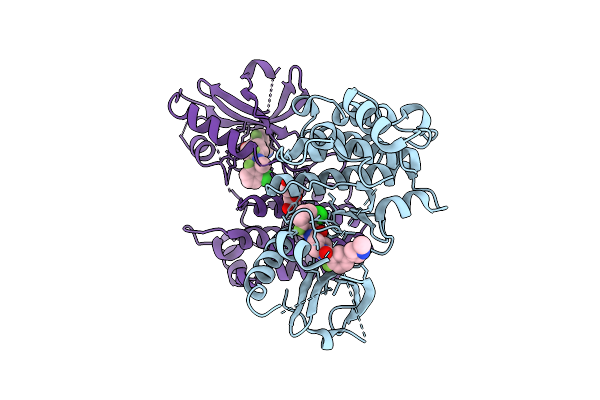 |
Crystal Structure Of An Allosteric Inhibitor Bound To Human Ripk1 Kinase Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: IMMUNE SYSTEM Ligands: A1IX9, IOD, PEG |
 |
Organism: Thermothielavioides terrestris (strain atcc 38088 / nrrl 8126)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: BIOSYNTHETIC PROTEIN Ligands: HEM, MG |
 |
Structural Studies On The Conformation Changes Induced By Ligand Binding In An Adenine Phosphoribosyltransferase (Fnaprt) From Fusobacterium Nucleatum
Organism: Fusobacterium nucleatum
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: TRANSFERASE Ligands: PO4, AMP |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE Ligands: PLP |
 |
Organism: Lacticaseibacillus rhamnosus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: LYASE |
 |
Organism: Danio rerio
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: HYDROLASE Ligands: NAG |
 |
Crystal Structure Of Rm014, A Macaque-Derived Hiv V2-Apex-Targeting Antibody From Apexgt6 Immunization
|
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN Ligands: Y01, A1JGQ |