Search Count: 3,957
 |
Organism: Pyrenophora tritici-repentis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2026-01-21 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2026-01-21 Classification: LIGASE Ligands: GOL, P4K |
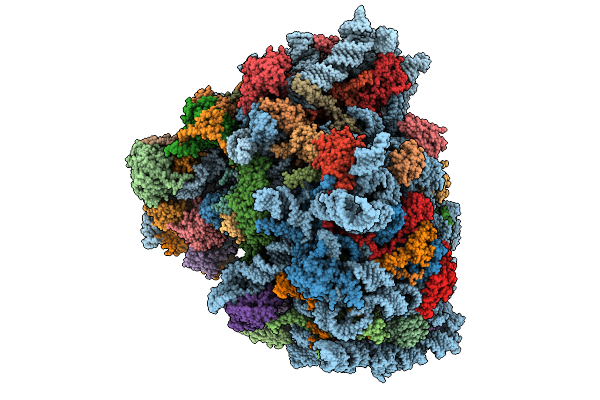 |
Rabbit 80S Ribosome In Complex With Erf1-Aaq, Stalled At The Stop Codon In Mutated F2A Sequence
Organism: Homo sapiens, Foot-and-mouth disease virus sat 2, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: RIBOSOME Ligands: MG, ZN, K, SPD, GTP, SF4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.79 Å Release Date: 2026-01-14 Classification: MOTOR PROTEIN Ligands: ADP, MG |
 |
Helical Reconstruction Of The Complex Of Pseudo-Acetylated Human Cardiac Actin (K326/328Q) And Tropomyosin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.12 Å Release Date: 2026-01-14 Classification: MOTOR PROTEIN Ligands: ADP, MG |
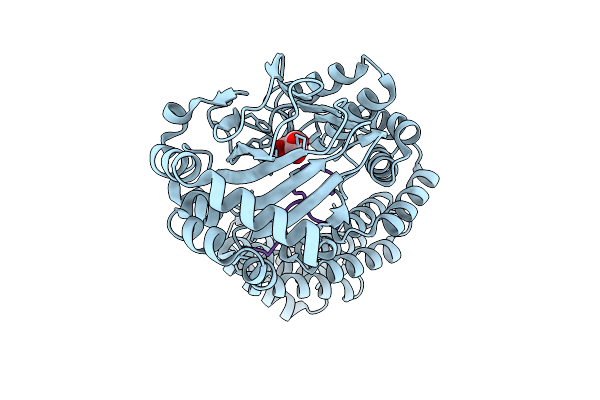 |
Room Temperature Structure Of Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate And Factor X Derived Peptide Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: AKG, FE |
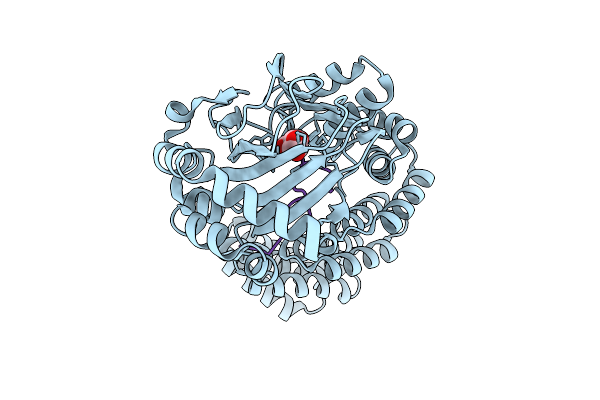 |
Room Temperature Structure Of Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate, And Hydroxylated Factor X Derived Peptide Fragment, 1.5 S O2 Exposure
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: AKG, SIN, FE |
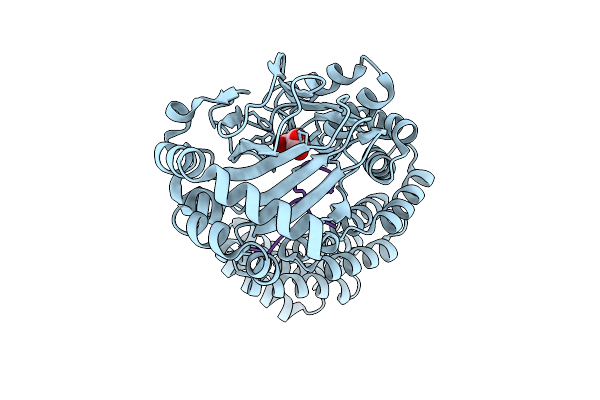 |
Room Temperature Structure Of Aspartyl/Asparaginyl Beta-Hydroxylase (Asph) In Complex With Fe, 2-Oxoglutarate And Factor X Derived Peptide Fragment, Excess Fe Removed
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2025-12-24 Classification: OXIDOREDUCTASE Ligands: AKG, FE |
 |
Organism: Enterococcus casseliflavus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-12-24 Classification: ANTIBIOTIC Ligands: A1I7W, DMS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: VIRAL PROTEIN/HYDROLASE |
 |
Cryo-Em Structure Of Active Human Green Cone Opsin In Complex With Chimeric G Protein (Minigist)
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: SIGNALING PROTEIN Ligands: RET |
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: MEMBRANE PROTEIN |
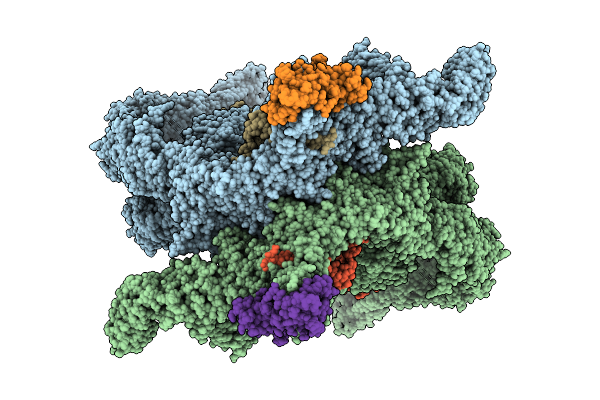 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-12-10 Classification: LIGASE Ligands: ZN |
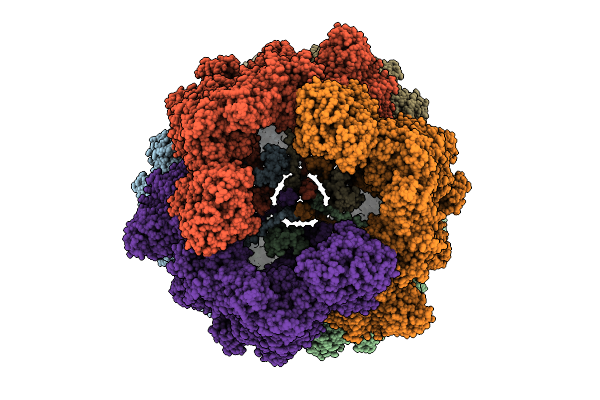 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN |
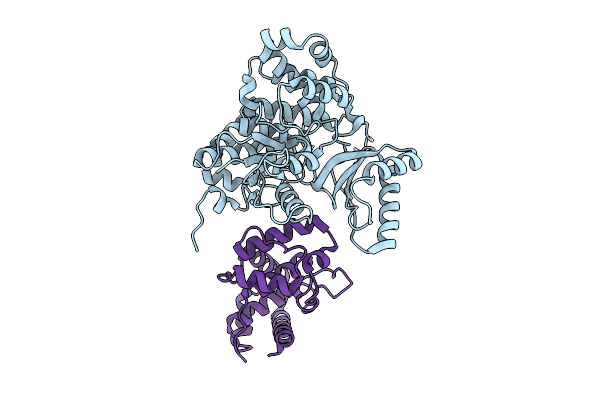 |
Structure Of Acp Domain Conjugated With Stearic Acid And Interacting With Mpt Domain From M. Tuberculosis Type-I Fas
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Resolution:3.51 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN |
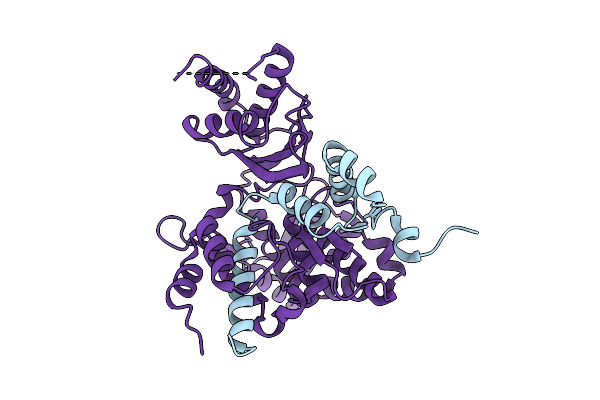 |
Structure Of Mpt Domain Of S. Cerevisiae Type-I Fas, Thio-Esterified To Palmitate
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:2.22 Å Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2025-12-03 Classification: SIGNALING PROTEIN |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-12-03 Classification: SIGNALING PROTEIN Ligands: MG |

