Search Count: 9,426
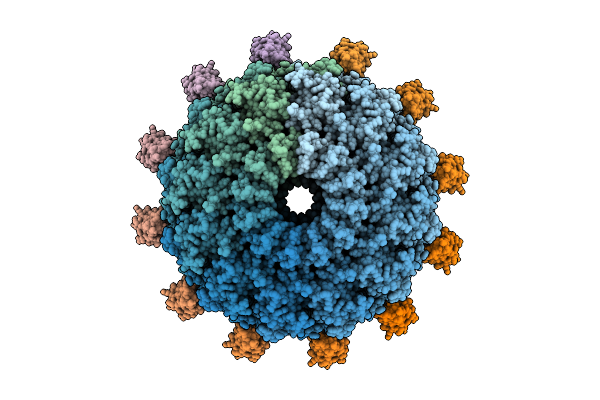 |
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS |
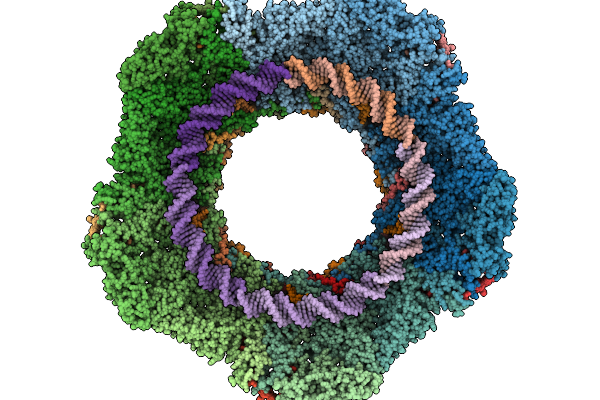 |
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS |
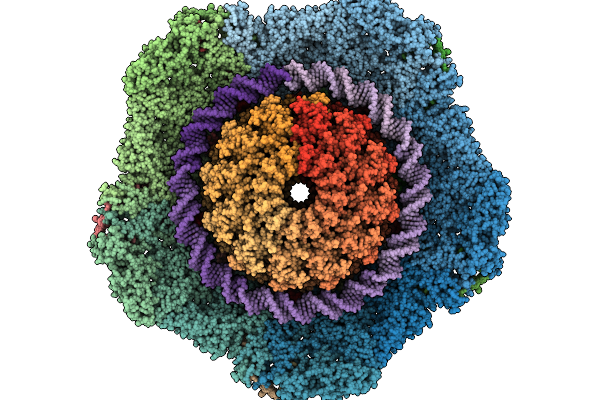 |
C1 Reconstruction Of The Thermophilic Bacteriophage P74-26 Portal And Portal Vertex
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS |
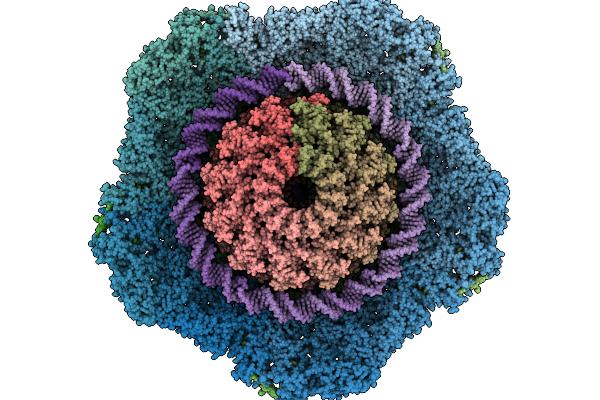 |
Composite Reconstruction Of The Thermophilic Bacteriophage P74-26 Neck And Portal Vertex
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS/DNA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2026-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1CKP, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2026-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1CKQ, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2026-01-28 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: A1CKS, SO4 |
 |
Organism: Oshimavirus p7426
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: VIRUS/DNA |
 |
Organism: Litorilinea aerophila
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: PROTEIN FIBRIL |
 |
Organism: Litorilinea aerophila
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: PROTEIN FIBRIL |
 |
Dihydrofolate Reductase Complexed With Folate And Nicotinamide Adenine Dinucleotide Phosphate (Oxidized Form)
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:0.89 Å Release Date: 2026-01-21 Classification: OXIDOREDUCTASE Ligands: NAP, FOL, MN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, IMD, A1JKS |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, IMD, EDO, CL, A1JMY |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1JMZ, IMD, EDO, CL, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1JM0, IMD, EDO, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: ZN, IMD, A1JM1 |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.73 Å Release Date: 2026-01-21 Classification: VIRAL PROTEIN Ligands: A1JM2, IMD, EDO, ZN |
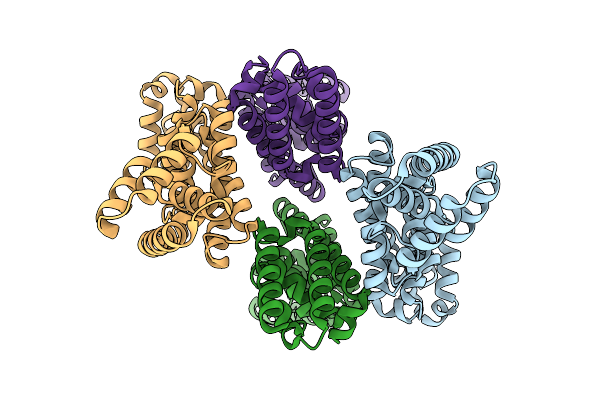 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Resolution:2.42 Å Release Date: 2026-01-21 Classification: PROTEIN TRANSPORT |
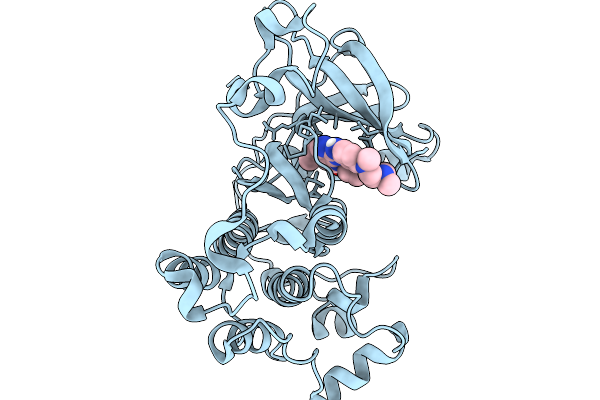 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: A1IR8 |
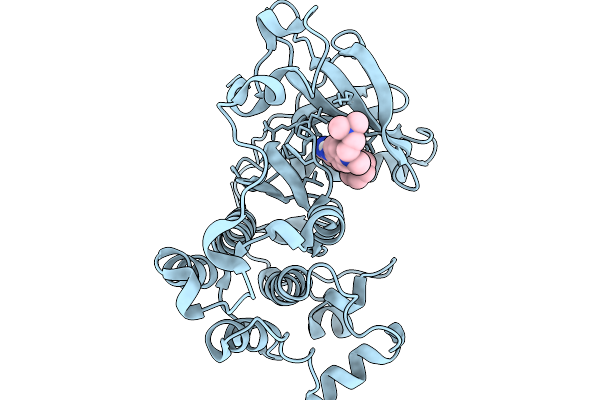 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2026-01-14 Classification: TRANSFERASE Ligands: A1IR9 |

