Search Count: 9,539
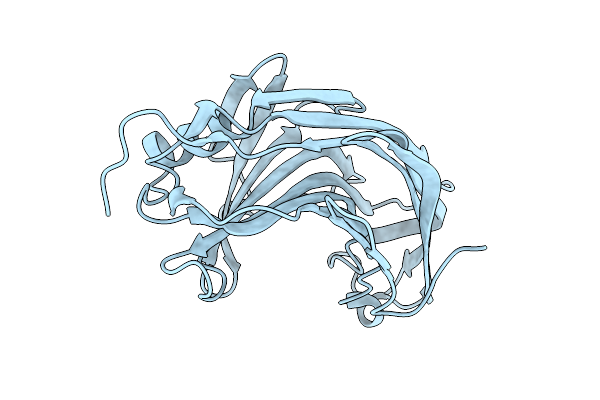 |
Crystal Structure Of The Pathogen-Secreted Apoplastic Gh12 Xyloglucan-Specific Endoglucanase Xeg1
Organism: Phytophthora sojae (strain p6497)
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: HYDROLASE |
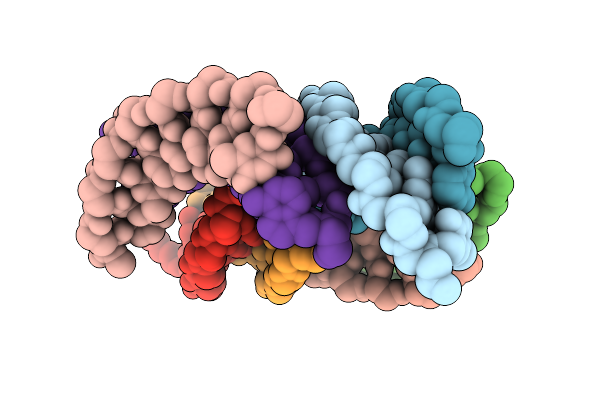 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: PEPTIDE NUCLEIC ACID |
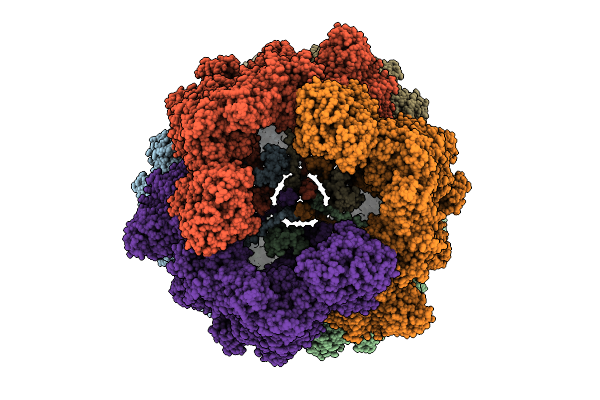 |
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN |
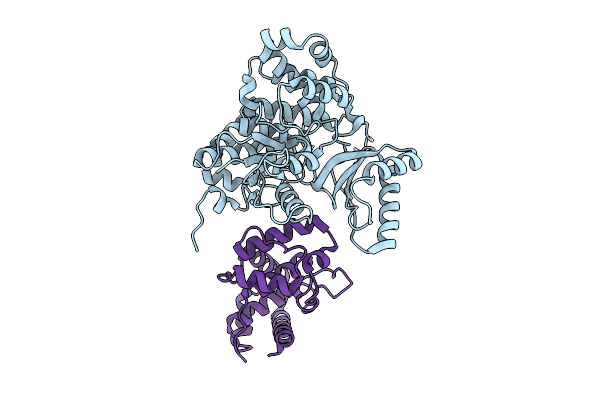 |
Structure Of Acp Domain Conjugated With Stearic Acid And Interacting With Mpt Domain From M. Tuberculosis Type-I Fas
Organism: Mycobacterium tuberculosis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Structure Of Mpt Domain Of S. Cerevisiae Type-I Fas, Thio-Esterified To Palmitate
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CELL CYCLE Ligands: GOL, SO4 |
 |
Cryo-Em Structure Of Lptdem Complex Containing Shigella Flexneri Lpte And Endogenous E. Coli Lptd And Lptm
Organism: Shigella flexneri, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde In Complex With Rtp45 Superinfection Exclusion Protein From Rtp Bacteriophage And Endogenous Lptm
Organism: Shigella flexneri, Escherichia phage rtp, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: PLM, PXS |
 |
Cryo-Em Structure Of The Open State Of Shigella Flexneri Lptde Bound By The Rbp Of Oekolampad Phage
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Dimer: Closed-State Unbound And Open-State Bound By Oekolampad Phage Rbp
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of Shigella Flexneri Lptde Bound By Phage Rbp Reveals N-Terminal Strand Insertion Into Lateral Gate
Organism: Shigella flexneri, Escherichia phage oekolampad
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of C278S Mutant Of Mouse Cdc14A In Complex With A Model Phosphopeptide
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: CELL CYCLE Ligands: GOL, SO4 |
 |
Organism: Mirabilis jalapa
Method: X-RAY DIFFRACTION Release Date: 2025-12-03 Classification: ANTIVIRAL PROTEIN Ligands: SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT Ligands: V6A |
 |
Cryo-Em Structure Of Human Exportin-1 Conjugated With Selinexor And Bound To Yeast Ran-Gtp And Human Asb8-Elob/C
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT Ligands: V6A, GTP, MG |
 |
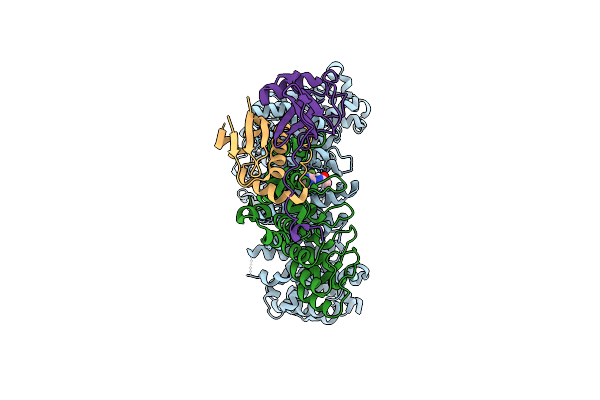
Cryo-Em Structure Of Human Exportin-1 Conjugated With Kpt-185 And Bound To Human Asb8-Elob/C
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT Ligands: K85 |
 |
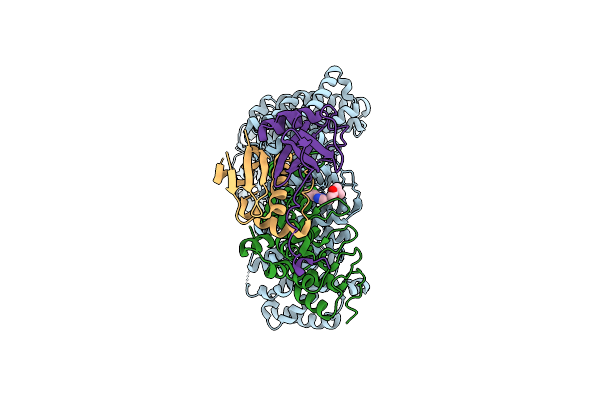
Cryo-Em Structure Of Human Exportin-1 Conjugated With Selinexor And Bound To Human Asb8(R197A)-Elob/C
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT Ligands: V6A |
 |
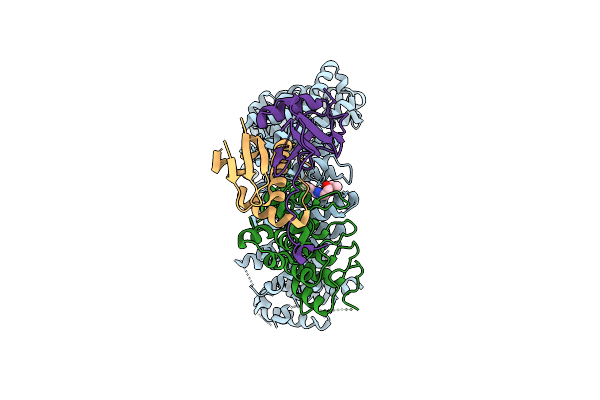
Cryo-Em Structure Of Human Exportin-1 Conjugated With Kpt-127 And Bound To Human Asb8-Elob/C
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT Ligands: A1CBC |
 |
Cryo-Em Structure Of Human Exportin-1 Conjugated With Kpt-Utsw1 And Bound To Human Asb8-Elob/C
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN TRANSPORT Ligands: A1CBB |

